+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
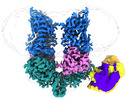
| Title | Structural Basis of Human NOX5 Activation | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  enzyme / enzyme /  oxidase / activation mechanism / oxidase / activation mechanism /  MEMBRANE PROTEIN MEMBRANE PROTEIN | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.99 Å cryo EM / Resolution: 3.99 Å | |||||||||
 Authors Authors | Cui C / Jiang M / Sun J | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structural Basis of Human NOX5 Activation Authors: Cui C / Jiang M / Sun J | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42348.map.gz emd_42348.map.gz | 49.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42348-v30.xml emd-42348-v30.xml emd-42348.xml emd-42348.xml | 12.2 KB 12.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_42348.png emd_42348.png | 78.5 KB | ||
| Filedesc metadata |  emd-42348.cif.gz emd-42348.cif.gz | 4.7 KB | ||
| Others |  emd_42348_half_map_1.map.gz emd_42348_half_map_1.map.gz emd_42348_half_map_2.map.gz emd_42348_half_map_2.map.gz | 48.8 MB 48.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42348 http://ftp.pdbj.org/pub/emdb/structures/EMD-42348 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42348 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42348 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42348.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42348.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.297 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_42348_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_42348_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : NADPH oxidase 5
| Entire | Name: NADPH oxidase 5 |
|---|---|
| Components |
|
-Supramolecule #1: NADPH oxidase 5
| Supramolecule | Name: NADPH oxidase 5 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: NADPH oxidase 5 (NOX5)
| Macromolecule | Name: NADPH oxidase 5 (NOX5) / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSAEEDARWL RWVTQQFKTI AGEDGEISLQ EFKAALHVKE SFFAERFFAL FDSDRSGTIT LQELQEALTL LIHGSPMDKL KFLFQVYDID GSGSIDPDEL RTVLQSCLRE SAISLPDEKL DQLTLALFES ADADGNGAIT FEELRDELQR FPGVMENLTI SAAHWLTAPA ...String: MSAEEDARWL RWVTQQFKTI AGEDGEISLQ EFKAALHVKE SFFAERFFAL FDSDRSGTIT LQELQEALTL LIHGSPMDKL KFLFQVYDID GSGSIDPDEL RTVLQSCLRE SAISLPDEKL DQLTLALFES ADADGNGAIT FEELRDELQR FPGVMENLTI SAAHWLTAPA PRPRPRRPRQ LTRAYWHNHR SQLFCLATYA GLHVLLFGLA ASAHRDLGAS VMVAKGCGQC LNFDCSFIAV LMLRRCLTWL RATWLAQVLP LDQNIQFHQL MGYVVVGLSL VHTVAHTVNF VLQAQAEASP FQFWELLLTT RPGIGWVHGS ASPTGVALLL LLLLMFICSS SCIRRSGHFE VFYWTHLSYL LVWLLLIFHG PNFWKWLLVP GILFFLEKAI GLAVSRMAAV CIMEVNLLPS KVTHLLIKRP PFFHYRPGDY LYLNIPTIAR YEWHPFTISS APEQKDTIWL HIRSQGQWTN RLYESFKASD PLGRGSKRLS RSVTMRKSQR SSKGSEILLE KHKFCNIKCY IDGPYGTPTR RIFASEHAVL IGAGIGITPF ASILQSIMYR HQKRKHTCPS CQHSWIEGVQ DNMKLHKVDF IWINRDQRSF EWFVSLLTKL EMDQAEEAQY GRFLELHMYM TSALGKNDMK AIGLQMALDL LANKEKKDSI TGLQTRTQPG RPDWSKVFQK VAAEKKGKVQ VFFCGSPALA KVLKGHCEKF GFRFFQENF |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.0 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.99 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 220000 |
 Movie
Movie Controller
Controller




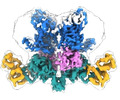
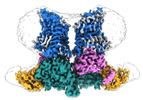
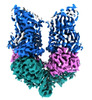





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)