[English] 日本語
 Yorodumi
Yorodumi- EMDB-41266: Cryo-EM structure of the Tripartite ATP-independent Periplasmic (... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Haemophilus influenzae (antiparallel dimer) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Sugar transport /  sialic acid / TRAP transporter / sialic acid / TRAP transporter /  secondary active transport / ion transporter superfamily / secondary active transport / ion transporter superfamily /  TRANSPORT PROTEIN TRANSPORT PROTEIN | |||||||||
| Function / homology | TRAP transporter large membrane protein DctM / TRAP transporter, small membrane protein DctQ / TRAP C4-dicarboxylate transport system permease DctM subunit / Tripartite ATP-independent periplasmic transporters, DctQ component /  Tripartite ATP-independent periplasmic transporter, DctM component / transmembrane transporter activity / carbohydrate transport / Tripartite ATP-independent periplasmic transporter, DctM component / transmembrane transporter activity / carbohydrate transport /  plasma membrane / Sialic acid TRAP transporter permease protein SiaT plasma membrane / Sialic acid TRAP transporter permease protein SiaT Function and homology information Function and homology information | |||||||||
| Biological species |   Haemophilus influenzae Rd KW20 (bacteria) Haemophilus influenzae Rd KW20 (bacteria) | |||||||||
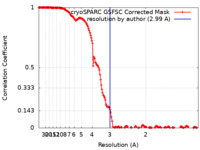
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.99 Å cryo EM / Resolution: 2.99 Å | |||||||||
 Authors Authors | Davies JS / Currie MC / Dobson RCJ / North RA | |||||||||
| Funding support |  New Zealand, 2 items New Zealand, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2024 Journal: Elife / Year: 2024Title: Structural and biophysical analysis of a tripartite ATP-independent periplasmic (TRAP) transporter. Authors: Michael J Currie / James S Davies / Mariafrancesca Scalise / Ashutosh Gulati / Joshua D Wright / Michael C Newton-Vesty / Gayan S Abeysekera / Ramaswamy Subramanian / Weixiao Y Wahlgren / ...Authors: Michael J Currie / James S Davies / Mariafrancesca Scalise / Ashutosh Gulati / Joshua D Wright / Michael C Newton-Vesty / Gayan S Abeysekera / Ramaswamy Subramanian / Weixiao Y Wahlgren / Rosmarie Friemann / Jane R Allison / Peter D Mace / Michael D W Griffin / Borries Demeler / Soichi Wakatsuki / David Drew / Cesare Indiveri / Renwick C J Dobson / Rachel A North /       Abstract: Tripartite ATP-independent periplasmic (TRAP) transporters are secondary-active transporters that receive their substrates via a soluble-binding protein to move bioorganic acids across bacterial or ...Tripartite ATP-independent periplasmic (TRAP) transporters are secondary-active transporters that receive their substrates via a soluble-binding protein to move bioorganic acids across bacterial or archaeal cell membranes. Recent cryo-electron microscopy (cryo-EM) structures of TRAP transporters provide a broad framework to understand how they work, but the mechanistic details of transport are not yet defined. Here we report the cryo-EM structure of the -acetylneuraminate TRAP transporter (SiaQM) at 2.99 Å resolution (extending to 2.2 Å at the core), revealing new features. The improved resolution (the previous SiaQM structure is 4.7 Å resolution) permits accurate assignment of two Na sites and the architecture of the substrate-binding site, consistent with mutagenic and functional data. Moreover, rather than a monomer, the SiaQM structure is a homodimer. We observe lipids at the dimer interface, as well as a lipid trapped within the fusion that links the SiaQ and SiaM subunits. We show that the affinity () for the complex between the soluble SiaP protein and SiaQM is in the micromolar range and that a related SiaP can bind SiaQM. This work provides key data that enhances our understanding of the 'elevator-with-an-operator' mechanism of TRAP transporters. #1:  Journal: Elife / Year: 2023 Journal: Elife / Year: 2023Title: Structural and biophysical analysis of a Haemophilus influenzae tripartite ATP-independent periplasmic (TRAP) transporter Authors: Currie MJ / Davies JS / Scalise M / Gulati A / Wright JD / Newton-Vesty MC / Abeysekera GS / Subramanian R / Wahlgren WY / Friemann R / Allison JR / Mace PD / Griffin MDW / Demeler B / ...Authors: Currie MJ / Davies JS / Scalise M / Gulati A / Wright JD / Newton-Vesty MC / Abeysekera GS / Subramanian R / Wahlgren WY / Friemann R / Allison JR / Mace PD / Griffin MDW / Demeler B / Wakatsuki S / Drew D / Indiveri C / Dobson RCJ / North RA | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41266.map.gz emd_41266.map.gz | 208.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41266-v30.xml emd-41266-v30.xml emd-41266.xml emd-41266.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41266_fsc.xml emd_41266_fsc.xml | 15.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_41266.png emd_41266.png | 111.2 KB | ||
| Filedesc metadata |  emd-41266.cif.gz emd-41266.cif.gz | 6.4 KB | ||
| Others |  emd_41266_half_map_1.map.gz emd_41266_half_map_1.map.gz emd_41266_half_map_2.map.gz emd_41266_half_map_2.map.gz | 390.6 MB 390.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41266 http://ftp.pdbj.org/pub/emdb/structures/EMD-41266 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41266 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41266 | HTTPS FTP |
-Related structure data
| Related structure data |  8thjMC  8thiC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_41266.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41266.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.6645 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_41266_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_41266_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : HiSiaQM transporter protein solubilised in amphipol A8-35
| Entire | Name: HiSiaQM transporter protein solubilised in amphipol A8-35 |
|---|---|
| Components |
|
-Supramolecule #1: HiSiaQM transporter protein solubilised in amphipol A8-35
| Supramolecule | Name: HiSiaQM transporter protein solubilised in amphipol A8-35 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Haemophilus influenzae Rd KW20 (bacteria) Haemophilus influenzae Rd KW20 (bacteria) |
-Macromolecule #1: Sialic acid TRAP transporter permease protein SiaT
| Macromolecule | Name: Sialic acid TRAP transporter permease protein SiaT / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Haemophilus influenzae Rd KW20 (bacteria) Haemophilus influenzae Rd KW20 (bacteria) |
| Molecular weight | Theoretical: 72.046812 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MGGSHHHHHH GMASMTGGQQ MGRDLYDDDD KDRWGSELEM KYINKLEEWL GGALFIAIFG ILIAQILSRQ VFHSPLIWSE ELAKLLFVY VGMLGISVAV RKQEHVFIDF LTNLMPEKIR KFTNTFVQLL VFICIFLFIH FGIRTFNGAS FPIDALGGIS E KWIFAALP ...String: MGGSHHHHHH GMASMTGGQQ MGRDLYDDDD KDRWGSELEM KYINKLEEWL GGALFIAIFG ILIAQILSRQ VFHSPLIWSE ELAKLLFVY VGMLGISVAV RKQEHVFIDF LTNLMPEKIR KFTNTFVQLL VFICIFLFIH FGIRTFNGAS FPIDALGGIS E KWIFAALP VVAILMMFRF IQAQTLNFKT GKSYLPATFF IISAVILFAI LFFAPDWFKV LRISNYIKLG SSSVYVALLV WL IIMFIGV PVGWSLFIAT LLYFSMTRWN VVNAATEKLV YSLDSFPLLA VPFYILTGIL MNTGGITERI FNFAKALLGH YTG GMGHVN IGASLLFSGM SGSALADAGG LGQLEIKAMR DAGYDDDICG GITAASCIIG PLVPPSIAMI IYGVIANESI AKLF IAGFI PGVLITLALM AMNYRIAKKR GYPRTPKATR EQLCSSFKQS FWAILTPLLI IGGIFSGLFS PTESAIVAAA YSVII GKFV YKELTLKSLF NSCIEAMAIT GVVALMIMTV TFFGDMIARE QVAMRVADVF VAVADSPLTV LIMINALLLF LGMFID ALA LQFLVLPMLI PIAMQFNIDL IFFGVMTTLN MMVGILTPPM GMALFVVARV GNMSVSTVTK GVLPFLIPVF VTLVLIT IF PQIITFVPNL LIP UniProtKB: Sialic acid TRAP transporter permease protein SiaT |
-Macromolecule #2: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 2 / Number of copies: 4 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #3: PHOSPHATIDYLETHANOLAMINE
| Macromolecule | Name: PHOSPHATIDYLETHANOLAMINE / type: ligand / ID: 3 / Number of copies: 2 / Formula: PTY |
|---|---|
| Molecular weight | Theoretical: 734.039 Da |
| Chemical component information |  ChemComp-PTY: |
-Macromolecule #4: (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-...
| Macromolecule | Name: (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE type: ligand / ID: 4 / Number of copies: 2 / Formula: PGT |
|---|---|
| Molecular weight | Theoretical: 751.023 Da |
| Chemical component information |  ChemComp-PGT: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.1 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.4 µm Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.4 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 73.1 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X