+ Open data
Open data
- Basic information
Basic information
| Entry | 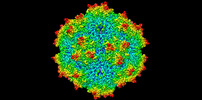 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Avian Adeno-associated virus - empty capsid | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Adeno-associated virus / Adeno-associated virus /  capsid / quail bronchitis / avianAAV / AAAV / capsid / quail bronchitis / avianAAV / AAAV /  VIRUS VIRUS | |||||||||
| Function / homology | Phospholipase A2-like domain / Phospholipase A2-like domain / Parvovirus coat protein VP2 / Parvovirus coat protein VP1/VP2 / Parvovirus coat protein VP2 / Capsid/spike protein, ssDNA virus / T=1 icosahedral viral capsid / structural molecule activity /  Capsid protein Capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |  Avian adeno-associated virus Avian adeno-associated virus | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.54 Å cryo EM / Resolution: 2.54 Å | |||||||||
 Authors Authors | Hsi J / Mietzsch M / Chipman P / Afione S / Zeher A / Huang R / Chiorini J / McKenna R | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Virol / Year: 2023 Journal: J Virol / Year: 2023Title: Structural and antigenic characterization of the avian adeno-associated virus capsid. Authors: Jane Hsi / Mario Mietzsch / Paul Chipman / Sandra Afione / Allison Zeher / Rick Huang / John Chiorini / Robert McKenna /  Abstract: AAVs are extensively studied as promising therapeutic gene delivery vectors. In order to circumvent pre-existing antibodies targeting primate-based AAV capsids, the AAAV capsid was evaluated as an ...AAVs are extensively studied as promising therapeutic gene delivery vectors. In order to circumvent pre-existing antibodies targeting primate-based AAV capsids, the AAAV capsid was evaluated as an alternative to primate-based therapeutic vectors. Despite the high sequence diversity, the AAAV capsid was found to bind to a common glycan receptor, terminal galactose, which is also utilized by other AAVs already being utilized in gene therapy trials. However, contrary to the initial hypothesis, AAAV was recognized by approximately 30% of human sera tested. Structural and sequence comparisons point to conserved epitopes in the fivefold region of the capsid as the reason determinant for the observed cross-reactivity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41208.map.gz emd_41208.map.gz | 262.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41208-v30.xml emd-41208-v30.xml emd-41208.xml emd-41208.xml | 14 KB 14 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41208.png emd_41208.png | 69.8 KB | ||
| Filedesc metadata |  emd-41208.cif.gz emd-41208.cif.gz | 5.4 KB | ||
| Others |  emd_41208_half_map_1.map.gz emd_41208_half_map_1.map.gz emd_41208_half_map_2.map.gz emd_41208_half_map_2.map.gz | 81 MB 81 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41208 http://ftp.pdbj.org/pub/emdb/structures/EMD-41208 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41208 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41208 | HTTPS FTP |
-Related structure data
| Related structure data |  8texMC  8teyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41208.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41208.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.052 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_41208_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_41208_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Avian adeno-associated virus
| Entire | Name:  Avian adeno-associated virus Avian adeno-associated virus |
|---|---|
| Components |
|
-Supramolecule #1: Avian adeno-associated virus
| Supramolecule | Name: Avian adeno-associated virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 341671 / Sci species name: Avian adeno-associated virus / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: Yes |
|---|
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 60 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Avian adeno-associated virus Avian adeno-associated virus |
| Molecular weight | Theoretical: 60.2375 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAEGGGGPVG DAGQGADGVG NSSGNWHCDS QWLENGVVTR TTRTWVLPSY NNHLYKRIQG PSGGDNNNKF FGFSTPWGYF DYNRFHCHF SPRDWQRLIN NNWGIRPKAM RFRLFNIQVK EVTVQDSNTT IANNLTSTVQ VFADKDYQLP YVLGSATEGT F PPFPADIY ...String: MAEGGGGPVG DAGQGADGVG NSSGNWHCDS QWLENGVVTR TTRTWVLPSY NNHLYKRIQG PSGGDNNNKF FGFSTPWGYF DYNRFHCHF SPRDWQRLIN NNWGIRPKAM RFRLFNIQVK EVTVQDSNTT IANNLTSTVQ VFADKDYQLP YVLGSATEGT F PPFPADIY TIPQYGYCTL NYNNEAVDRS AFYCLDYFPS DMLRTGNNFE FTYTFEDVPF HSMFAHNQTL DRLMNPLVDQ YL WAFSSVS QAGSSGRALH YSRATKTNMA AQYRNWLPGP FFRDQQIFTG ASNITKNNVF SVWEKGKQWE LDNRTNLMQP GPA AATTFS GEPDRQAMQN TLAFSRTVYD QTTATTDRNQ ILITNEDEIR PTNSVGIDAW GAVPTNNQSI VTPGTRAAVN NQGA LPGMV WQNRDIYLQG PIWAKIPDTD NHFHPSPLIG GFGCKHPPPQ IFIKNTPVPA NPSETFQTAK VASFINQYST GQCTV EIFW ELKKETSKRW NPEIQFTSNF GNAADIQFAV SDTGSYSEPR PIGTRYLTKP L UniProtKB:  Capsid protein Capsid protein |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.4 µm Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.4 µm |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 61.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Initial angle assignment | Type: COMMON LINE |
| Final angle assignment | Type: COMMON LINE |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.54 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 38672 |
 Movie
Movie Controller
Controller







 Z
Z Y
Y X
X

















