[English] 日本語
 Yorodumi
Yorodumi- EMDB-41180: Global reconstruction for HCMV Pentamer in complex with CS2pt1p2_... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Global reconstruction for HCMV Pentamer in complex with CS2pt1p2_A10L Fab and CS3pt1p4_C1L Fab | |||||||||
 Map data Map data | Global reconstruction half map A for HCMV Pentamer in complex with CS2pt1p2_A10L Fab and CS3pt1p4_C1L Fab | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Virus / Virus /  glycoprotein / glycoprotein /  antibody / antibody /  VIRAL PROTEIN VIRAL PROTEIN | |||||||||
| Biological species |    Human betaherpesvirus 5 / Human betaherpesvirus 5 /   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.4 Å cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Goldsmith JG / McLellan JS | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Immunity / Year: 2023 Journal: Immunity / Year: 2023Title: Single-cell analysis of memory B cells from top neutralizers reveals multiple sites of vulnerability within HCMV Trimer and Pentamer. Authors: Matthias Zehner / Mira Alt / Artem Ashurov / Jory A Goldsmith / Rebecca Spies / Nina Weiler / Justin Lerma / Lutz Gieselmann / Dagmar Stöhr / Henning Gruell / Eric P Schultz / Christoph ...Authors: Matthias Zehner / Mira Alt / Artem Ashurov / Jory A Goldsmith / Rebecca Spies / Nina Weiler / Justin Lerma / Lutz Gieselmann / Dagmar Stöhr / Henning Gruell / Eric P Schultz / Christoph Kreer / Linda Schlachter / Hanna Janicki / Kerstin Laib Sampaio / Cora Stegmann / Michelle D Nemetchek / Sabrina Dähling / Leon Ullrich / Ulf Dittmer / Oliver Witzke / Manuel Koch / Brent J Ryckman / Ramin Lotfi / Jason S McLellan / Adalbert Krawczyk / Christian Sinzger / Florian Klein /   Abstract: Human cytomegalovirus (HCMV) can cause severe diseases in fetuses, newborns, and immunocompromised individuals. Currently, no vaccines are approved, and treatment options are limited. Here, we ...Human cytomegalovirus (HCMV) can cause severe diseases in fetuses, newborns, and immunocompromised individuals. Currently, no vaccines are approved, and treatment options are limited. Here, we analyzed the human B cell response of four HCMV top neutralizers from a cohort of 9,000 individuals. By single-cell analyses of memory B cells targeting the pentameric and trimeric HCMV surface complexes, we identified vulnerable sites on the shared gH/gL subunits as well as complex-specific subunits UL and gO. Using high-resolution cryogenic electron microscopy, we revealed the structural basis of the neutralization mechanisms of antibodies targeting various binding sites. Moreover, we identified highly potent antibodies that neutralized a broad spectrum of HCMV strains, including primary clinical isolates, that outperform known antibodies used in clinical trials. Our study provides a deep understanding of the mechanisms of HCMV neutralization and identifies promising antibody candidates to prevent and treat HCMV infection. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41180.map.gz emd_41180.map.gz | 122.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41180-v30.xml emd-41180-v30.xml emd-41180.xml emd-41180.xml | 16.5 KB 16.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41180.png emd_41180.png | 41.1 KB | ||
| Filedesc metadata |  emd-41180.cif.gz emd-41180.cif.gz | 4.1 KB | ||
| Others |  emd_41180_additional_1.map.gz emd_41180_additional_1.map.gz emd_41180_half_map_1.map.gz emd_41180_half_map_1.map.gz emd_41180_half_map_2.map.gz emd_41180_half_map_2.map.gz | 214.6 MB 226.4 MB 226.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41180 http://ftp.pdbj.org/pub/emdb/structures/EMD-41180 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41180 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41180 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_41180.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41180.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Global reconstruction half map A for HCMV Pentamer in complex with CS2pt1p2_A10L Fab and CS3pt1p4_C1L Fab | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.94 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Global reconstruction half sharpened map for HCMV Pentamer...
| File | emd_41180_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Global reconstruction half sharpened map for HCMV Pentamer in complex with CS2pt1p2_A10L Fab and CS3pt1p4_C1L Fab | ||||||||||||
| Projections & Slices |
| ||||||||||||
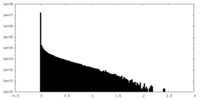
| Density Histograms |
-Half map: Global reconstruction map for HCMV Pentamer in complex...
| File | emd_41180_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Global reconstruction map for HCMV Pentamer in complex with CS2pt1p2_A10L Fab and CS3pt1p4_C1L Fab | ||||||||||||
| Projections & Slices |
| ||||||||||||
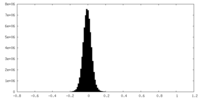
| Density Histograms |
-Half map: Global reconstruction half map B for HCMV Pentamer...
| File | emd_41180_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Global reconstruction half map B for HCMV Pentamer in complex with CS2pt1p2_A10L Fab and CS3pt1p4_C1L Fab | ||||||||||||
| Projections & Slices |
| ||||||||||||
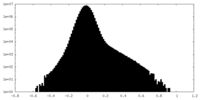
| Density Histograms |
- Sample components
Sample components
-Entire : HCMV Pentamer in complex with CS2pt1p2_A10L Fab and CS3pt1p4_C1L Fab
| Entire | Name: HCMV Pentamer in complex with CS2pt1p2_A10L Fab and CS3pt1p4_C1L Fab |
|---|---|
| Components |
|
-Supramolecule #1: HCMV Pentamer in complex with CS2pt1p2_A10L Fab and CS3pt1p4_C1L Fab
| Supramolecule | Name: HCMV Pentamer in complex with CS2pt1p2_A10L Fab and CS3pt1p4_C1L Fab type: complex / ID: 1 / Parent: 0 |
|---|
-Supramolecule #2: HCMV Pentamer
| Supramolecule | Name: HCMV Pentamer / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:    Human betaherpesvirus 5 Human betaherpesvirus 5 |
-Supramolecule #3: CS2pt1p2_A10L Fab and CS3pt1p4_C1L Fab
| Supramolecule | Name: CS2pt1p2_A10L Fab and CS3pt1p4_C1L Fab / type: complex / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm Bright-field microscopy / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 49.0 e/Å2 |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 71677 |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)