+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a full-length, native Drp1 dimer | |||||||||
 Map data Map data | Drp1 cryoEM native dimer map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Dynamin-related protein 1 /  Drp1 / Drp1 /  mitochondrial fission / mitochondrial fission /  GTPase / GTPase /  Cytosolic Protein Cytosolic Protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationmitochondrial membrane fission / regulation of ATP metabolic process / regulation of peroxisome organization / mitocytosis / Apoptotic execution phase /  dynamin GTPase / peroxisome fission / dynamin GTPase / peroxisome fission /  regulation of mitophagy / mitochondrial fragmentation involved in apoptotic process / GTP-dependent protein binding ...mitochondrial membrane fission / regulation of ATP metabolic process / regulation of peroxisome organization / mitocytosis / Apoptotic execution phase / regulation of mitophagy / mitochondrial fragmentation involved in apoptotic process / GTP-dependent protein binding ...mitochondrial membrane fission / regulation of ATP metabolic process / regulation of peroxisome organization / mitocytosis / Apoptotic execution phase /  dynamin GTPase / peroxisome fission / dynamin GTPase / peroxisome fission /  regulation of mitophagy / mitochondrial fragmentation involved in apoptotic process / GTP-dependent protein binding / protein localization to mitochondrion / regulation of mitophagy / mitochondrial fragmentation involved in apoptotic process / GTP-dependent protein binding / protein localization to mitochondrion /  mitochondrial fission / positive regulation of neutrophil chemotaxis / regulation of mitochondrion organization / positive regulation of mitochondrial fission / intracellular distribution of mitochondria / mitochondrial fission / positive regulation of neutrophil chemotaxis / regulation of mitochondrion organization / positive regulation of mitochondrial fission / intracellular distribution of mitochondria /  heart contraction / necroptotic process / positive regulation of release of cytochrome c from mitochondria / heart contraction / necroptotic process / positive regulation of release of cytochrome c from mitochondria /  brush border / localization / positive regulation of intrinsic apoptotic signaling pathway / brush border / localization / positive regulation of intrinsic apoptotic signaling pathway /  clathrin-coated pit / clathrin-coated pit /  GTPase activator activity / mitochondrion organization / release of cytochrome c from mitochondria / positive regulation of protein secretion / synaptic vesicle membrane / GTPase activator activity / mitochondrion organization / release of cytochrome c from mitochondria / positive regulation of protein secretion / synaptic vesicle membrane /  small GTPase binding / small GTPase binding /  peroxisome / peroxisome /  endocytosis / calcium ion transport / rhythmic process / protein complex oligomerization / endocytosis / calcium ion transport / rhythmic process / protein complex oligomerization /  microtubule binding / microtubule binding /  regulation of gene expression / protein-containing complex assembly / regulation of gene expression / protein-containing complex assembly /  microtubule / mitochondrial outer membrane / microtubule / mitochondrial outer membrane /  membrane fusion / molecular adaptor activity / positive regulation of apoptotic process / intracellular membrane-bounded organelle / membrane fusion / molecular adaptor activity / positive regulation of apoptotic process / intracellular membrane-bounded organelle /  GTPase activity / GTPase activity /  lipid binding / lipid binding /  ubiquitin protein ligase binding / endoplasmic reticulum membrane / GTP binding / perinuclear region of cytoplasm / ubiquitin protein ligase binding / endoplasmic reticulum membrane / GTP binding / perinuclear region of cytoplasm /  Golgi apparatus / Golgi apparatus /  endoplasmic reticulum / protein homodimerization activity / protein-containing complex / endoplasmic reticulum / protein homodimerization activity / protein-containing complex /  mitochondrion / mitochondrion /  membrane / identical protein binding / membrane / identical protein binding /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
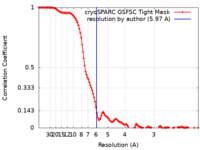
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 5.97 Å cryo EM / Resolution: 5.97 Å | |||||||||
 Authors Authors | Rochon K / Mears JA | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural basis for regulated assembly of the mitochondrial fission GTPase Drp1. Authors: Kristy Rochon / Brianna L Bauer / Nathaniel A Roethler / Yuli Buckley / Chih-Chia Su / Wei Huang / Rajesh Ramachandran / Maria S K Stoll / Edward W Yu / Derek J Taylor / Jason A Mears /  Abstract: Mitochondrial fission is a critical cellular event to maintain organelle function. This multistep process is initiated by the enhanced recruitment and oligomerization of dynamin-related protein 1 ...Mitochondrial fission is a critical cellular event to maintain organelle function. This multistep process is initiated by the enhanced recruitment and oligomerization of dynamin-related protein 1 (Drp1) at the surface of mitochondria. As such, Drp1 is essential for inducing mitochondrial division in mammalian cells, and homologous proteins are found in all eukaryotes. As a member of the dynamin superfamily of proteins (DSPs), controlled Drp1 self-assembly into large helical polymers stimulates its GTPase activity to promote membrane constriction. Still, little is known about the mechanisms that regulate correct spatial and temporal assembly of the fission machinery. Here we present a cryo-EM structure of a full-length Drp1 dimer in an auto-inhibited state. This dimer reveals two key conformational rearrangements that must be unlocked through intramolecular rearrangements to achieve the assembly-competent state observed in previous structures. This structural insight provides understanding into the mechanism for regulated self-assembly of the mitochondrial fission machinery. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40967.map.gz emd_40967.map.gz | 156.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40967-v30.xml emd-40967-v30.xml emd-40967.xml emd-40967.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40967_fsc.xml emd_40967_fsc.xml | 11.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_40967.png emd_40967.png | 47.9 KB | ||
| Filedesc metadata |  emd-40967.cif.gz emd-40967.cif.gz | 6.5 KB | ||
| Others |  emd_40967_half_map_1.map.gz emd_40967_half_map_1.map.gz emd_40967_half_map_2.map.gz emd_40967_half_map_2.map.gz | 154.2 MB 154.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40967 http://ftp.pdbj.org/pub/emdb/structures/EMD-40967 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40967 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40967 | HTTPS FTP |
-Related structure data
| Related structure data |  8t1hMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40967.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40967.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Drp1 cryoEM native dimer map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Drp1 cryoEM native dimer half map B
| File | emd_40967_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Drp1 cryoEM native dimer half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Drp1 cryoEM native dimer half map A
| File | emd_40967_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Drp1 cryoEM native dimer half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Dimer complex of native, full length Drp1
| Entire | Name: Dimer complex of native, full length Drp1 |
|---|---|
| Components |
|
-Supramolecule #1: Dimer complex of native, full length Drp1
| Supramolecule | Name: Dimer complex of native, full length Drp1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 164 KDa |
-Macromolecule #1: Dynamin-1-like protein
| Macromolecule | Name: Dynamin-1-like protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number:  dynamin GTPase dynamin GTPase |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 81.984094 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MEALIPVINK LQDVFNTVGA DIIQLPQIVV VGTQSSGKSS VLESLVGRDL LPRGTGIVTR RPLILQLVHV SQEDKRKTTG EENGVEAEE WGKFLHTKNK LYTDFDEIRQ EIENETERIS GNNKGVSPEP IHLKIFSPNV VNLTLVDLPG MTKVPVGDQP K DIELQIRE ...String: MEALIPVINK LQDVFNTVGA DIIQLPQIVV VGTQSSGKSS VLESLVGRDL LPRGTGIVTR RPLILQLVHV SQEDKRKTTG EENGVEAEE WGKFLHTKNK LYTDFDEIRQ EIENETERIS GNNKGVSPEP IHLKIFSPNV VNLTLVDLPG MTKVPVGDQP K DIELQIRE LILRFISNPN SIILAVTAAN TDMATSEALK ISREVDPDGR RTLAVITKLD LMDAGTDAMD VLMGRVIPVK LG IIGVVNR SQLDINNKKS VTDSIRDEYA FLQKKYPSLA NRNGTKYLAR TLNRLLMHHI RDCLPELKTR INVLAAQYQS LLN SYGEPV DDKSATLLQL ITKFATEYCN TIEGTAKYIE TSELCGGARI CYIFHETFGR TLESVDPLGG LNTIDILTAI RNAT GPRPA LFVPEVSFEL LVKRQIKRLE EPSLRCVELV HEEMQRIIQH CSNYSTQELL RFPKLHDAIV EVVTCLLRKR LPVTN EMVH NLVAIELAYI NTKHPDFADA CGLMNNNIEE QRRNRLAREL PSAVSRDKSS KVPSALAPAS QEPSPAASAE ADGKLI QDS RRETKNVASG GGGVGDGVQE PTTGNWRGML KTSKAEELLA EEKSKPIPIM PASPQKGHAV NLLDVPVPVA RKLSARE QR DCEVIERLIK SYFLIVRKNI QDSVPKAVMH FLVNHVKDTL QSELVGQLYK SSLLDDLLTE SEDMAQRRKE AADMLKAL Q GASQIIAEIR ETHLW UniProtKB: Dynamin-1-like protein |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | .05 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: 25 mM HEPES (KOH) pH 7.5, 0.15 M KCl, 5 mM MgCl2, 10 mM Beta-mercaptoethanol | |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number real images: 4560 / Average electron dose: 47.76 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)