[English] 日本語
 Yorodumi
Yorodumi- EMDB-40406: In situ structure of Helicobacter pylori flagellar motor from Pil... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | In situ structure of Helicobacter pylori flagellar motor from PilN and PilO deletion mutant | |||||||||
 Map data Map data | A subtomogram averaged structure of Helicobacter pylori flagellar motor from PilN and PilO deletion mutant | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | H.pylori /  Flagella / PilN / PilO / Flagella / PilN / PilO /  MOTOR PROTEIN MOTOR PROTEIN | |||||||||
| Biological species |   Helicobacter pylori G27 (bacteria) Helicobacter pylori G27 (bacteria) | |||||||||
| Method | subtomogram averaging /  cryo EM / Resolution: 39.0 Å cryo EM / Resolution: 39.0 Å | |||||||||
 Authors Authors | Tachiyama S / Liu J | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: Bacterial flagella hijack type IV pili proteins to control motility. Authors: Xiaolin Liu / Shoichi Tachiyama / Xiaotian Zhou / Rommel A Mathias / Sharmin Q Bonny / Mohammad F Khan / Yue Xin / Anna Roujeinikova / Jun Liu / Karen M Ottemann /   Abstract: Bacterial flagella and type IV pili (TFP) are surface appendages that enable motility and mechanosensing through distinct mechanisms. These structures were previously thought to have no components in ...Bacterial flagella and type IV pili (TFP) are surface appendages that enable motility and mechanosensing through distinct mechanisms. These structures were previously thought to have no components in common. Here, we report that TFP and some flagella share proteins PilO, PilN, and PilM, which we identified as part of the flagellar motor. mutants lacking PilO or PilN migrated better than wild type in semisolid agar because they continued swimming rather than aggregated into microcolonies, mimicking the TFP-regulated surface response. Like their TFP homologs, flagellar PilO/PilN heterodimers formed a peripheral cage that encircled the flagellar motor. These results indicate that PilO and PilN act similarly in flagella and TFP by differentially regulating motility and microcolony formation when bacteria encounter surfaces. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40406.map.gz emd_40406.map.gz | 6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40406-v30.xml emd-40406-v30.xml emd-40406.xml emd-40406.xml | 13.7 KB 13.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_40406.png emd_40406.png | 79.6 KB | ||
| Filedesc metadata |  emd-40406.cif.gz emd-40406.cif.gz | 4.2 KB | ||
| Others |  emd_40406_half_map_1.map.gz emd_40406_half_map_1.map.gz emd_40406_half_map_2.map.gz emd_40406_half_map_2.map.gz | 6 MB 6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40406 http://ftp.pdbj.org/pub/emdb/structures/EMD-40406 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40406 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40406 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40406.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40406.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | A subtomogram averaged structure of Helicobacter pylori flagellar motor from PilN and PilO deletion mutant | ||||||||||||||||||||
| Voxel size | X=Y=Z: 8.592 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: An EM half map of Helicobacter pylori flagellar...
| File | emd_40406_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
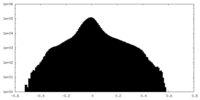
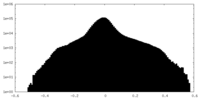
| Annotation | An EM half map of Helicobacter pylori flagellar motor from PilN and PilO deletion mutant | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: An EM half map of Helicobacter pylori flagellar...
| File | emd_40406_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
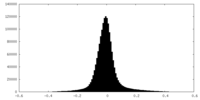
| Annotation | An EM half map of Helicobacter pylori flagellar motor from PilN and PilO deletion mutant | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Helicobacter pylori
| Entire | Name:   Helicobacter pylori (bacteria) Helicobacter pylori (bacteria) |
|---|---|
| Components |
|
-Supramolecule #1: Helicobacter pylori
| Supramolecule | Name: Helicobacter pylori / type: cell / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:   Helicobacter pylori G27 (bacteria) Helicobacter pylori G27 (bacteria) |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 5.0 µm / Nominal defocus min: 3.0 µm Bright-field microscopy / Nominal defocus max: 5.0 µm / Nominal defocus min: 3.0 µm |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 1.81 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Extraction | Number tomograms: 166 / Number images used: 478 |
|---|---|
| Final angle assignment | Type: OTHER |
| Final reconstruction | Applied symmetry - Point group: C18 (18 fold cyclic ) / Resolution.type: BY AUTHOR / Resolution: 39.0 Å / Resolution method: FSC 0.5 CUT-OFF / Number subtomograms used: 6928 ) / Resolution.type: BY AUTHOR / Resolution: 39.0 Å / Resolution method: FSC 0.5 CUT-OFF / Number subtomograms used: 6928 |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X