[English] 日本語
 Yorodumi
Yorodumi- EMDB-40194: The Type 9 Secretion System Extended Translocon - SprA-PorV-PPI-R... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
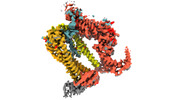
| Title | The Type 9 Secretion System Extended Translocon - SprA-PorV-PPI-RemZ-SkpA-SprE complex | |||||||||||||||||||||
 Map data Map data | Composite volume used for refinement of coordinates created from EMD-29911 and EMD-40085 the associated consensus volume is EMD-40086 | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | T9SS / Type IX Secretion System Translocon / Protein Secretion PorV - SprA - PPI - RemZ Complex /  MEMBRANE PROTEIN MEMBRANE PROTEIN | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology information peptidylprolyl isomerase / peptidylprolyl isomerase /  peptidyl-prolyl cis-trans isomerase activity / unfolded protein binding peptidyl-prolyl cis-trans isomerase activity / unfolded protein bindingSimilarity search - Function | |||||||||||||||||||||
| Biological species |  Flavobacterium johnsoniae UW101 (bacteria) / Flavobacterium johnsoniae UW101 (bacteria) /  Flavobacterium johnsoniae (bacteria) Flavobacterium johnsoniae (bacteria) | |||||||||||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.2 Å cryo EM / Resolution: 2.2 Å | |||||||||||||||||||||
 Authors Authors | Deme JC / Lea SM | |||||||||||||||||||||
| Funding support |  United Kingdom, European Union, United Kingdom, European Union,  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2024 Journal: Nat Microbiol / Year: 2024Title: The Type 9 Secretion System caught in the act of transport Authors: Lauber F / Deme JC / Liu X / Kjaer A / Miller HL / Alcock F / Lea SM / Berks BC | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40194.map.gz emd_40194.map.gz | 42.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40194-v30.xml emd-40194-v30.xml emd-40194.xml emd-40194.xml | 21.8 KB 21.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_40194.png emd_40194.png | 47 KB | ||
| Filedesc metadata |  emd-40194.cif.gz emd-40194.cif.gz | 9.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40194 http://ftp.pdbj.org/pub/emdb/structures/EMD-40194 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40194 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40194 | HTTPS FTP |
-Related structure data
| Related structure data |  8gl8MC  8gl6C  8gljC  8glkC  8glmC  8glnC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40194.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40194.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite volume used for refinement of coordinates created from EMD-29911 and EMD-40085 the associated consensus volume is EMD-40086 | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
+Entire : Type 9 Secretion System Extended Translocon
+Supramolecule #1: Type 9 Secretion System Extended Translocon
+Macromolecule #1: Protein involved in gliding motility SprA
+Macromolecule #2: Peptidyl-prolyl cis-trans isomerase
+Macromolecule #3: Type IX secretion system protein PorV domain-containing protein
+Macromolecule #4: RemZ
+Macromolecule #5: Periplasmic chaperone for outer membrane proteins Skp
+Macromolecule #6: SprE
+Macromolecule #7: Lauryl Maltose Neopentyl Glycol
+Macromolecule #8: alpha-D-mannopyranose
+Macromolecule #9: water
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.2 µm Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.2 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 61.2 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 3.0) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 3.0) |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.2 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.0) / Number images used: 546490 |
 Movie
Movie Controller
Controller