[English] 日本語
 Yorodumi
Yorodumi- EMDB-40047: Structure of human ENPP1 in complex with variable heavy domain VH27.2 -
+ Open data
Open data
- Basic information
Basic information
| Entry | 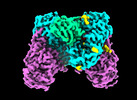 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human ENPP1 in complex with variable heavy domain VH27.2 | |||||||||
 Map data Map data | From cryosparc non-uniform refinement and local resolution filter | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  phosphodiesterase / phosphodiesterase /  inhibitor / inhibitor /  IMMUNE SYSTEM IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationGTP diphosphatase activity / cyclic-GMP-AMP hydrolase activity / negative regulation of hh target transcription factor activity / inorganic diphosphate transport / Vitamin B2 (riboflavin) metabolism / UTP diphosphatase activity /  phosphodiesterase I / 3'-phosphoadenosine 5'-phosphosulfate binding / dinucleotide phosphatase activity / Vitamin B5 (pantothenate) metabolism ...GTP diphosphatase activity / cyclic-GMP-AMP hydrolase activity / negative regulation of hh target transcription factor activity / inorganic diphosphate transport / Vitamin B2 (riboflavin) metabolism / UTP diphosphatase activity / phosphodiesterase I / 3'-phosphoadenosine 5'-phosphosulfate binding / dinucleotide phosphatase activity / Vitamin B5 (pantothenate) metabolism ...GTP diphosphatase activity / cyclic-GMP-AMP hydrolase activity / negative regulation of hh target transcription factor activity / inorganic diphosphate transport / Vitamin B2 (riboflavin) metabolism / UTP diphosphatase activity /  phosphodiesterase I / 3'-phosphoadenosine 5'-phosphosulfate binding / dinucleotide phosphatase activity / Vitamin B5 (pantothenate) metabolism / nucleoside triphosphate catabolic process / phosphodiesterase I / 3'-phosphoadenosine 5'-phosphosulfate binding / dinucleotide phosphatase activity / Vitamin B5 (pantothenate) metabolism / nucleoside triphosphate catabolic process /  nucleotide diphosphatase / 3'-phosphoadenosine 5'-phosphosulfate metabolic process / intracellular phosphate ion homeostasis / negative regulation of protein autophosphorylation / nucleoside triphosphate diphosphatase activity / nucleic acid metabolic process / sequestering of triglyceride / negative regulation of bone mineralization / nucleotide diphosphatase / 3'-phosphoadenosine 5'-phosphosulfate metabolic process / intracellular phosphate ion homeostasis / negative regulation of protein autophosphorylation / nucleoside triphosphate diphosphatase activity / nucleic acid metabolic process / sequestering of triglyceride / negative regulation of bone mineralization /  ATP diphosphatase activity / negative regulation of glycogen biosynthetic process / phosphate ion homeostasis / melanocyte differentiation / ATP diphosphatase activity / negative regulation of glycogen biosynthetic process / phosphate ion homeostasis / melanocyte differentiation /  phosphodiesterase I activity / scavenger receptor activity / negative regulation of glucose import / phosphodiesterase I activity / scavenger receptor activity / negative regulation of glucose import /  regulation of bone mineralization / phosphate-containing compound metabolic process / regulation of bone mineralization / phosphate-containing compound metabolic process /  exonuclease activity / negative regulation of fat cell differentiation / exonuclease activity / negative regulation of fat cell differentiation /  polysaccharide binding / response to ATP / polysaccharide binding / response to ATP /  bone mineralization / bone mineralization /  immunoglobulin complex / immunoglobulin complex /  phosphatase activity / phosphatase activity /  3',5'-cyclic-AMP phosphodiesterase activity / response to inorganic substance / ATP metabolic process / negative regulation of insulin receptor signaling pathway / generation of precursor metabolites and energy / 3',5'-cyclic-AMP phosphodiesterase activity / response to inorganic substance / ATP metabolic process / negative regulation of insulin receptor signaling pathway / generation of precursor metabolites and energy /  insulin receptor binding / negative regulation of cell growth / cellular response to insulin stimulus / insulin receptor binding / negative regulation of cell growth / cellular response to insulin stimulus /  gene expression / basolateral plasma membrane / gene expression / basolateral plasma membrane /  adaptive immune response / adaptive immune response /  nucleic acid binding / nucleic acid binding /  immune response / lysosomal membrane / immune response / lysosomal membrane /  calcium ion binding / calcium ion binding /  cell surface / protein homodimerization activity / cell surface / protein homodimerization activity /  extracellular space / zinc ion binding / extracellular region / extracellular space / zinc ion binding / extracellular region /  ATP binding / ATP binding /  membrane / membrane /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
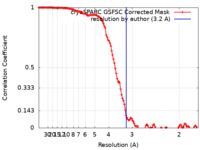
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.2 Å cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Carozza JA / Wang H / Solomon PE / Wells JA / Li L | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2024 Journal: Nat Chem Biol / Year: 2024Title: Discovery of VH domains that allosterically inhibit ENPP1. Authors: Paige E Solomon / Colton J Bracken / Jacqueline A Carozza / Haoqing Wang / Elizabeth P Young / Alon Wellner / Chang C Liu / E Alejandro Sweet-Cordero / Lingyin Li / James A Wells /  Abstract: Ectodomain phosphatase/phosphodiesterase-1 (ENPP1) is overexpressed on cancer cells and functions as an innate immune checkpoint by hydrolyzing extracellular cyclic guanosine monophosphate adenosine ...Ectodomain phosphatase/phosphodiesterase-1 (ENPP1) is overexpressed on cancer cells and functions as an innate immune checkpoint by hydrolyzing extracellular cyclic guanosine monophosphate adenosine monophosphate (cGAMP). Biologic inhibitors have not yet been reported and could have substantial therapeutic advantages over current small molecules because they can be recombinantly engineered into multifunctional formats and immunotherapies. Here we used phage and yeast display coupled with in cellulo evolution to generate variable heavy (VH) single-domain antibodies against ENPP1 and discovered a VH domain that allosterically inhibited the hydrolysis of cGAMP and adenosine triphosphate (ATP). We solved a 3.2 Å-resolution cryo-electron microscopy structure for the VH inhibitor complexed with ENPP1 that confirmed its new allosteric binding pose. Finally, we engineered the VH domain into multispecific formats and immunotherapies, including a bispecific fusion with an anti-PD-L1 checkpoint inhibitor that showed potent cellular activity. | |||||||||
| History |
|
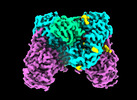
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40047.map.gz emd_40047.map.gz | 5.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40047-v30.xml emd-40047-v30.xml emd-40047.xml emd-40047.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40047_fsc.xml emd_40047_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_40047.png emd_40047.png | 93.1 KB | ||
| Filedesc metadata |  emd-40047.cif.gz emd-40047.cif.gz | 6.4 KB | ||
| Others |  emd_40047_half_map_1.map.gz emd_40047_half_map_1.map.gz emd_40047_half_map_2.map.gz emd_40047_half_map_2.map.gz | 165.1 MB 165.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40047 http://ftp.pdbj.org/pub/emdb/structures/EMD-40047 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40047 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40047 | HTTPS FTP |
-Related structure data
| Related structure data |  8ghrMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40047.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40047.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | From cryosparc non-uniform refinement and local resolution filter | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8677 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map A from non-uniform refinement
| File | emd_40047_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A from non-uniform refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B from non-uniform refinement
| File | emd_40047_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B from non-uniform refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ENPP1-VH27 complex
| Entire | Name: ENPP1-VH27 complex |
|---|---|
| Components |
|
-Supramolecule #1: ENPP1-VH27 complex
| Supramolecule | Name: ENPP1-VH27 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Ectonucleotide pyrophosphatase/phosphodiesterase family member 1,...
| Macromolecule | Name: Ectonucleotide pyrophosphatase/phosphodiesterase family member 1, secreted form, Immunoglobulin gamma-1 heavy chain fusion type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 115.488617 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GEKSWVEEPC ESINEPQCPA GFETPPTLLF SLDGFRAEYL HTWGGLLPVI SKLKKCGTYT KNMRPVYPTK AFPNHYSIVT GLYPESHGI IDNKMYDPKM NASFSLKSKE KFNPEWYKGE PIWVTAKYQG LKSGTFFWPG SDVEINGIFP DIYKMYNGSV P FEERILAV ...String: GEKSWVEEPC ESINEPQCPA GFETPPTLLF SLDGFRAEYL HTWGGLLPVI SKLKKCGTYT KNMRPVYPTK AFPNHYSIVT GLYPESHGI IDNKMYDPKM NASFSLKSKE KFNPEWYKGE PIWVTAKYQG LKSGTFFWPG SDVEINGIFP DIYKMYNGSV P FEERILAV LQWLQLPKDE RPHFYTLYLE EPDSSGHSYG PVSSEVIKAL QRVDGMVGML MDGLKELNLH RCLNLILISD HG MEQGSCK KYIYLNKYLG DVKNIKVIYG PAARLRPSDV PDKYYSFNYE GIARNLSCRE PNQHFKPYLK HFLPKRLHFA KSD RIEPLT FYLDPQWQLA LNPSERKYCG SGFHGSDNVF SNMQALFVGY GPGFKHGIEA DTFENIEVYN LMCDLLNLTP APNN GTHGS LNHLLKNPVY TPKHPKEVHP LVQCPFTRNP RDNLGCSCNP SILPIEDFQT QFNLTVAEEK IIKHETLPYG RPRVL QKEN TICLLSQHQF MSGYSQDILM PLWTSYTVDR NDSFSTEDFS NCLYQDFRIP LSPVHKCSFY KNNTKVSYGF LSPPQL NKN SSGIYSEALL TTNIVPMYQS FQVIWRYFHD TLLRKYAEER NGVNVVSGPV FDFDYDGRCD SLENLRQKRR VIRNQEI LI PTHFFIVLTS CKDTSQTPLH CENLDTLAFI LPHRTDNSES CVHGKHDSSW VEELLMLHRA RITDVEHITG LSFYQQRK E PVSDILKLKT HLPTFSQEDT SSGGGGENLY FQSSGGGSGG GEPKSCDKTH TCPPCPAPEL LGGPSVFLFP PKPKDTLMI SRTPEVTCVV VDVSHEDPEV KFNWYVDGVE VHNAKTKPRE EQYNSTYRVV SVLTVLHQDW LNGKEYKCKV SNKALPAPIE KTISKAKGQ PREPQVYTLP PSRDELTKNQ VSLTCLVKGF YPSDIAVEWE SNGQPENNYK TTPPVLDSDG SFFLYSKLTV D KSRWQQGN VFSCSVMHEA LHNHYTQKSL SLSPGKGGGG SGLNDIFEAQ KIEWHEG UniProtKB: Ectonucleotide pyrophosphatase/phosphodiesterase family member 1, Immunoglobulin gamma-1 heavy chain |
-Macromolecule #2: VH27
| Macromolecule | Name: VH27 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.564035 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGGG LVQPGGSLRL SCAASGFDIY YSYYIGWVRR APGKGEELVA RISPSYGSTS YADSVKGRFT ISADISKNTA YLQMNSLRV EDTAVYYCAR FAYPWYVADD ALDYWGQGTL VTVSS |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #5: ADENOSINE MONOPHOSPHATE
| Macromolecule | Name: ADENOSINE MONOPHOSPHATE / type: ligand / ID: 5 / Number of copies: 2 / Formula: AMP |
|---|---|
| Molecular weight | Theoretical: 347.221 Da |
| Chemical component information |  ChemComp-AMP: |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 4 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #7: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 7 / Number of copies: 2 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 6 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: phosphate buffered saline |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.7000000000000001 µm Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.7000000000000001 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 57.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z
Z Y
Y X
X