[English] 日本語
 Yorodumi
Yorodumi- EMDB-38134: The cryo-EM structure of insect gustatory receptor Gr43a I418A fr... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The cryo-EM structure of insect gustatory receptor Gr43a I418A from Drosophila melanogaster in complex with fructose | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | gustatory receptor /  ligand-gated ion channel / Gr43a / ligand-gated ion channel / Gr43a /  fructose / fructose /  MEMBRANE PROTEIN MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsweet taste receptor activity /  taste receptor activity / male courtship behavior / taste receptor activity / male courtship behavior /  regulation of appetite / adult feeding behavior / chemosensory behavior / sensory perception of sweet taste / regulation of appetite / adult feeding behavior / chemosensory behavior / sensory perception of sweet taste /  regulation of feeding behavior / cellular response to fructose stimulus / sensory perception of taste ...sweet taste receptor activity / regulation of feeding behavior / cellular response to fructose stimulus / sensory perception of taste ...sweet taste receptor activity /  taste receptor activity / male courtship behavior / taste receptor activity / male courtship behavior /  regulation of appetite / adult feeding behavior / chemosensory behavior / sensory perception of sweet taste / regulation of appetite / adult feeding behavior / chemosensory behavior / sensory perception of sweet taste /  regulation of feeding behavior / cellular response to fructose stimulus / sensory perception of taste / regulation of feeding behavior / cellular response to fructose stimulus / sensory perception of taste /  axon / axon /  dendrite / neuronal cell body / dendrite / neuronal cell body /  signal transduction / signal transduction /  membrane / membrane /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Drosophila melanogaster (fruit fly) Drosophila melanogaster (fruit fly) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.8 Å cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Ma D / Guo J | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2024 Journal: Science / Year: 2024Title: Structural basis for sugar perception by gustatory receptors. Authors: Demin Ma / Meiqin Hu / Xiaotong Yang / Qiang Liu / Fan Ye / Weijie Cai / Yong Wang / Ximing Xu / Shenghai Chang / Ruiying Wang / Wei Yang / Sheng Ye / Nannan Su / Minrui Fan / Haoxing Xu / Jiangtao Guo /   Abstract: Insects rely on a family of seven transmembrane proteins called gustatory receptors (GRs) to encode different taste modalities, such as sweet and bitter. We report structures of sweet taste ...Insects rely on a family of seven transmembrane proteins called gustatory receptors (GRs) to encode different taste modalities, such as sweet and bitter. We report structures of sweet taste receptors GR43a and GR64a in the apo and sugar-bound states. Both GRs form tetrameric sugar-gated cation channels composed of one central pore domain (PD) and four peripheral ligand-binding domains (LBDs). Whereas GR43a is specifically activated by the monosaccharide fructose that binds to a narrow pocket in LBDs, disaccharides sucrose and maltose selectively activate GR64a by binding to a larger and flatter pocket in LBDs. Sugar binding to LBDs induces local conformational changes, which are subsequently transferred to the PD to cause channel opening. Our studies reveal a structural basis for sugar recognition and activation of GRs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38134.map.gz emd_38134.map.gz | 48.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38134-v30.xml emd-38134-v30.xml emd-38134.xml emd-38134.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_38134.png emd_38134.png | 42.2 KB | ||
| Filedesc metadata |  emd-38134.cif.gz emd-38134.cif.gz | 5.8 KB | ||
| Others |  emd_38134_half_map_1.map.gz emd_38134_half_map_1.map.gz emd_38134_half_map_2.map.gz emd_38134_half_map_2.map.gz | 45.1 MB 44.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38134 http://ftp.pdbj.org/pub/emdb/structures/EMD-38134 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38134 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38134 | HTTPS FTP |
-Related structure data
| Related structure data |  8x83MC  8jm9C  8jmaC  8jmeC  8jmhC  8jmiC  8x82C  8x84C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_38134.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38134.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.93 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_38134_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
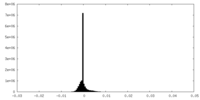
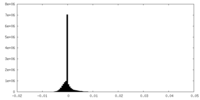
| Density Histograms |
-Half map: #1
| File | emd_38134_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
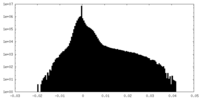
| Density Histograms |
- Sample components
Sample components
-Entire : gustatory receptor Gr43a I418A
| Entire | Name: gustatory receptor Gr43a I418A |
|---|---|
| Components |
|
-Supramolecule #1: gustatory receptor Gr43a I418A
| Supramolecule | Name: gustatory receptor Gr43a I418A / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Drosophila melanogaster (fruit fly) Drosophila melanogaster (fruit fly) |
-Macromolecule #1: Gustatory receptor for sugar taste 43a
| Macromolecule | Name: Gustatory receptor for sugar taste 43a / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Drosophila melanogaster (fruit fly) Drosophila melanogaster (fruit fly) |
| Molecular weight | Theoretical: 49.267312 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEISQPSIGI FYISKVLALA PYATVRNSKG RVEIGRSWLF TVYSATLTVV MVFLTYRGLL FDANSEIPVR MKSATSKVVT ALDVSVVVM AIVSGVYCGL FSLNDTLELN DRLNKIDNTL NAYNNFRRDR WRALGMAAVS LLAISILVGL DVGTWMRIAQ D MNIAQSDT ...String: MEISQPSIGI FYISKVLALA PYATVRNSKG RVEIGRSWLF TVYSATLTVV MVFLTYRGLL FDANSEIPVR MKSATSKVVT ALDVSVVVM AIVSGVYCGL FSLNDTLELN DRLNKIDNTL NAYNNFRRDR WRALGMAAVS LLAISILVGL DVGTWMRIAQ D MNIAQSDT ELNVHWYIPF YSLYFILTGL QVNIANTAYG LGRRFGRLNR MLSSSFLAEN NATSAIKPQK VSTVKNVSVN RP AMPSALH ASLTKLNGET LPSEAAAKNK GLLLKSLADS HESLGKCVHL LSNSFGIAVL FILVSCLLHL VATAYFLFLE LLS KRDNGY LWVQMLWICF HFLRLLMVVE PCHLAARESR KTIQIVCEIE RKVHEPILAE AVKKFWQQLL VVDADFSACG LCRV NRTIL TSFASAIATY LVALIQFQRT NGLEGGSSGG WSHPQFEK UniProtKB: Gustatory receptor for sugar taste 43a |
-Macromolecule #2: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 2 / Number of copies: 2 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #3: beta-D-fructofuranose
| Macromolecule | Name: beta-D-fructofuranose / type: ligand / ID: 3 / Number of copies: 4 / Formula: FRU |
|---|---|
| Molecular weight | Theoretical: 180.156 Da |
| Chemical component information | 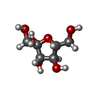 ChemComp-FRU: |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 4 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm Bright-field microscopy / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 52.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 158079 |
 Movie
Movie Controller
Controller










 Z
Z Y
Y X
X