[English] 日本語
 Yorodumi
Yorodumi- EMDB-37325: Cryo-EM structure of Escherichia coli Str K12 FtsE(E163Q)X/EnvC c... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Escherichia coli Str K12 FtsE(E163Q)X/EnvC complex with ATP in peptidisc | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  complex / complex /  TRANSPORT PROTEIN TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationdivision septum /  divisome complex / peptidoglycan-based cell wall biogenesis / Gram-negative-bacterium-type cell wall / septum digestion after cytokinesis / peptidoglycan turnover / divisome complex / peptidoglycan-based cell wall biogenesis / Gram-negative-bacterium-type cell wall / septum digestion after cytokinesis / peptidoglycan turnover /  plasma membrane protein complex / FtsZ-dependent cytokinesis / division septum assembly / cell division site ...division septum / plasma membrane protein complex / FtsZ-dependent cytokinesis / division septum assembly / cell division site ...division septum /  divisome complex / peptidoglycan-based cell wall biogenesis / Gram-negative-bacterium-type cell wall / septum digestion after cytokinesis / peptidoglycan turnover / divisome complex / peptidoglycan-based cell wall biogenesis / Gram-negative-bacterium-type cell wall / septum digestion after cytokinesis / peptidoglycan turnover /  plasma membrane protein complex / FtsZ-dependent cytokinesis / division septum assembly / cell division site / plasma membrane protein complex / FtsZ-dependent cytokinesis / division septum assembly / cell division site /  extrinsic component of membrane / extrinsic component of membrane /  ATPase complex / positive regulation of cell division / transmembrane transporter activity / response to radiation / transmembrane transport / ATPase complex / positive regulation of cell division / transmembrane transporter activity / response to radiation / transmembrane transport /  metalloendopeptidase activity / outer membrane-bounded periplasmic space / metalloendopeptidase activity / outer membrane-bounded periplasmic space /  periplasmic space / periplasmic space /  hydrolase activity / response to xenobiotic stimulus / hydrolase activity / response to xenobiotic stimulus /  cell cycle / cell cycle /  cell division / response to antibiotic / cell division / response to antibiotic /  ATP hydrolysis activity / ATP hydrolysis activity /  ATP binding / ATP binding /  plasma membrane / plasma membrane /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Escherichia coli K-12 (bacteria) Escherichia coli K-12 (bacteria) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.4 Å cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Li J / Xu X / He Y / Luo M | |||||||||
| Funding support |  Singapore, 1 items Singapore, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of Escherichia coli Str K12 FtsE(E163Q)X/EnvC complex with ATP in peptidisc Authors: Li J / Xu X / He Y / Luo M | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37325.map.gz emd_37325.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37325-v30.xml emd-37325-v30.xml emd-37325.xml emd-37325.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
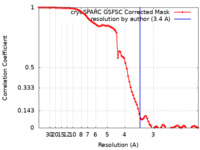
| FSC (resolution estimation) |  emd_37325_fsc.xml emd_37325_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_37325.png emd_37325.png | 31.4 KB | ||
| Filedesc metadata |  emd-37325.cif.gz emd-37325.cif.gz | 5.8 KB | ||
| Others |  emd_37325_half_map_1.map.gz emd_37325_half_map_1.map.gz emd_37325_half_map_2.map.gz emd_37325_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37325 http://ftp.pdbj.org/pub/emdb/structures/EMD-37325 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37325 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37325 | HTTPS FTP |
-Related structure data
| Related structure data |  8w6jMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37325.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37325.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_37325_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_37325_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ATP bound FtsE(E163Q)X-EnvC complex
| Entire | Name: ATP bound FtsE(E163Q)X-EnvC complex |
|---|---|
| Components |
|
-Supramolecule #1: ATP bound FtsE(E163Q)X-EnvC complex
| Supramolecule | Name: ATP bound FtsE(E163Q)X-EnvC complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Escherichia coli K-12 (bacteria) Escherichia coli K-12 (bacteria) |
-Macromolecule #1: Cell division ATP-binding protein FtsE
| Macromolecule | Name: Cell division ATP-binding protein FtsE / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli K-12 (bacteria) Escherichia coli K-12 (bacteria) |
| Molecular weight | Theoretical: 24.475295 KDa |
| Recombinant expression | Organism:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) |
| Sequence | String: MIRFEHVSKA YLGGRQALQG VTFHMQPGEM AFLTGHSGAG KSTLLKLICG IERPSAGKIW FSGHDITRLK NREVPFLRRQ IGMIFQDHH LLMDRTVYDN VAIPLIIAGA SGDDIRRRVS AALDKVGLLD KAKNFPIQLS GGEQQRVGIA RAVVNKPAVL L ADQPTGNL ...String: MIRFEHVSKA YLGGRQALQG VTFHMQPGEM AFLTGHSGAG KSTLLKLICG IERPSAGKIW FSGHDITRLK NREVPFLRRQ IGMIFQDHH LLMDRTVYDN VAIPLIIAGA SGDDIRRRVS AALDKVGLLD KAKNFPIQLS GGEQQRVGIA RAVVNKPAVL L ADQPTGNL DDALSEGILR LFEEFNRVGV TVLMATHDIN LISRRSYRML TLSDGHLHGG VGHE UniProtKB: Cell division ATP-binding protein FtsE |
-Macromolecule #2: Cell division protein FtsX
| Macromolecule | Name: Cell division protein FtsX / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli K-12 (bacteria) Escherichia coli K-12 (bacteria) |
| Molecular weight | Theoretical: 38.5835 KDa |
| Recombinant expression | Organism:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) |
| Sequence | String: MNKRDAINHI RQFGGRLDRF RKSVGGSGDG GRNAPKRAKS SPKPVNRKTN VFNEQVRYAF HGALQDLKSK PFATFLTVMV IAISLTLPS VCYMVYKNVN QAATQYYPSP QITVYLQKTL DDDAAAGVVA QLQAEQGVEK VNYLSREDAL GEFRNWSGFG G ALDMLEEN ...String: MNKRDAINHI RQFGGRLDRF RKSVGGSGDG GRNAPKRAKS SPKPVNRKTN VFNEQVRYAF HGALQDLKSK PFATFLTVMV IAISLTLPS VCYMVYKNVN QAATQYYPSP QITVYLQKTL DDDAAAGVVA QLQAEQGVEK VNYLSREDAL GEFRNWSGFG G ALDMLEEN PLPAVAVVIP KLDFQGTESL NTLRDRITQI NGIDEVRMDD SWFARLAALT GLVGRVSAMI GVLMVAAVFL VI GNSVRLS IFARRDSINV QKLIGATDGF ILRPFLYGGA LLGFSGALLS LILSEILVLR LSSAVAEVAQ VFGTKFDING LSF DECLLL LLVCSMIGWV AAWLATVQHL RHFTPE UniProtKB:  Cell division protein FtsX Cell division protein FtsX |
-Macromolecule #3: Murein hydrolase activator EnvC
| Macromolecule | Name: Murein hydrolase activator EnvC / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli K-12 (bacteria) Escherichia coli K-12 (bacteria) |
| Molecular weight | Theoretical: 46.661617 KDa |
| Recombinant expression | Organism:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) |
| Sequence | String: MTRAVKPRRF AIRPIIYASV LSAGVLLCAF SAHADERDQL KSIQADIAAK ERAVRQKQQQ RASLLAQLKK QEEAISEATR KLRETQNTL NQLNKQIDEM NASIAKLEQQ KAAQERSLAA QLDAAFRQGE HTGIQLILSG EESQRGQRLQ AYFGYLNQAR Q ETIAQLKQ ...String: MTRAVKPRRF AIRPIIYASV LSAGVLLCAF SAHADERDQL KSIQADIAAK ERAVRQKQQQ RASLLAQLKK QEEAISEATR KLRETQNTL NQLNKQIDEM NASIAKLEQQ KAAQERSLAA QLDAAFRQGE HTGIQLILSG EESQRGQRLQ AYFGYLNQAR Q ETIAQLKQ TREEVAMQRA ELEEKQSEQQ TLLYEQRAQQ AKLTQALNER KKTLAGLESS IQQGQQQLSE LRANESRLRN SI ARAEAAA KARAEREARE AQAVRDRQKE ATRKGTTYKP TESEKSLMSR TGGLGAPRGQ AFWPVRGPTL HRYGEQLQGE LRW KGMVIG ASEGTEVKAI ADGRVILADW LQGYGLVVVV EHGKGDMSLY GYNQSALVSV GSQVRAGQPI ALVGSSGGQG RPSL YFEIR RQGQAVNPQP WLGR UniProtKB: Murein hydrolase activator EnvC |
-Macromolecule #4: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 2.5 µm / Calibrated defocus min: 1.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Average exposure time: 6.02 sec. / Average electron dose: 38.837 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z
Z Y
Y X
X