[English] 日本語
 Yorodumi
Yorodumi- EMDB-36478: Focused refiment structure of NTSR1 in NTSR1-GRK2-Galpha(q) complexes -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Focused refiment structure of NTSR1 in NTSR1-GRK2-Galpha(q) complexes | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Biased signaling /  SIGNALING PROTEIN SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationG protein-coupled neurotensin receptor activity / inositol phosphate catabolic process / positive regulation of inhibitory postsynaptic potential / symmetric synapse / D-aspartate import across plasma membrane / positive regulation of gamma-aminobutyric acid secretion / L-glutamate import across plasma membrane / positive regulation of arachidonic acid secretion / regulation of respiratory gaseous exchange / negative regulation of release of sequestered calcium ion into cytosol ...G protein-coupled neurotensin receptor activity / inositol phosphate catabolic process / positive regulation of inhibitory postsynaptic potential / symmetric synapse / D-aspartate import across plasma membrane / positive regulation of gamma-aminobutyric acid secretion / L-glutamate import across plasma membrane / positive regulation of arachidonic acid secretion / regulation of respiratory gaseous exchange / negative regulation of release of sequestered calcium ion into cytosol / negative regulation of systemic arterial blood pressure / positive regulation of glutamate secretion / positive regulation of inositol phosphate biosynthetic process /  temperature homeostasis / response to lipid / temperature homeostasis / response to lipid /  regulation of membrane depolarization / detection of temperature stimulus involved in sensory perception of pain / neuropeptide signaling pathway / Peptide ligand-binding receptors / adult locomotory behavior / positive regulation of release of sequestered calcium ion into cytosol / dendritic shaft / regulation of membrane depolarization / detection of temperature stimulus involved in sensory perception of pain / neuropeptide signaling pathway / Peptide ligand-binding receptors / adult locomotory behavior / positive regulation of release of sequestered calcium ion into cytosol / dendritic shaft /  learning / G protein-coupled receptor activity / learning / G protein-coupled receptor activity /  terminal bouton / cytoplasmic side of plasma membrane / terminal bouton / cytoplasmic side of plasma membrane /  perikaryon / chemical synaptic transmission / G alpha (q) signalling events / perikaryon / chemical synaptic transmission / G alpha (q) signalling events /  dendritic spine / positive regulation of apoptotic process / dendritic spine / positive regulation of apoptotic process /  membrane raft / G protein-coupled receptor signaling pathway / protein-containing complex binding / positive regulation of gene expression / negative regulation of apoptotic process / membrane raft / G protein-coupled receptor signaling pathway / protein-containing complex binding / positive regulation of gene expression / negative regulation of apoptotic process /  Golgi apparatus / Golgi apparatus /  cell surface / cell surface /  endoplasmic reticulum / identical protein binding / endoplasmic reticulum / identical protein binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.02 Å cryo EM / Resolution: 3.02 Å | |||||||||
 Authors Authors | Duan J / Liu H / Zhao F / Yuan Q / Ji Y / Xu HE | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: GPCR activation and GRK2 assembly by a biased intracellular agonist. Authors: Jia Duan / Heng Liu / Fenghui Zhao / Qingning Yuan / Yujie Ji / Xiaoqing Cai / Xinheng He / Xinzhu Li / Junrui Li / Kai Wu / Tianyu Gao / Shengnan Zhu / Shi Lin / Ming-Wei Wang / Xi Cheng / ...Authors: Jia Duan / Heng Liu / Fenghui Zhao / Qingning Yuan / Yujie Ji / Xiaoqing Cai / Xinheng He / Xinzhu Li / Junrui Li / Kai Wu / Tianyu Gao / Shengnan Zhu / Shi Lin / Ming-Wei Wang / Xi Cheng / Wanchao Yin / Yi Jiang / Dehua Yang / H Eric Xu /  Abstract: Phosphorylation of G-protein-coupled receptors (GPCRs) by GPCR kinases (GRKs) desensitizes G-protein signalling and promotes arrestin signalling, which is also modulated by biased ligands. The ...Phosphorylation of G-protein-coupled receptors (GPCRs) by GPCR kinases (GRKs) desensitizes G-protein signalling and promotes arrestin signalling, which is also modulated by biased ligands. The molecular assembly of GRKs on GPCRs and the basis of GRK-mediated biased signalling remain largely unknown owing to the weak GPCR-GRK interactions. Here we report the complex structure of neurotensin receptor 1 (NTSR1) bound to GRK2, Gα and the arrestin-biased ligand SBI-553. The density map reveals the arrangement of the intact GRK2 with the receptor, with the N-terminal helix of GRK2 docking into the open cytoplasmic pocket formed by the outward movement of the receptor transmembrane helix 6, analogous to the binding of the G protein to the receptor. SBI-553 binds at the interface between GRK2 and NTSR1 to enhance GRK2 binding. The binding mode of SBI-553 is compatible with arrestin binding but clashes with the binding of Gα protein, thus providing a mechanism for its arrestin-biased signalling capability. In sum, our structure provides a rational model for understanding the details of GPCR-GRK interactions and GRK2-mediated biased signalling. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36478.map.gz emd_36478.map.gz | 118 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36478-v30.xml emd-36478-v30.xml emd-36478.xml emd-36478.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36478.png emd_36478.png | 26.1 KB | ||
| Masks |  emd_36478_msk_1.map emd_36478_msk_1.map | 125 MB |  Mask map Mask map | |
| Others |  emd_36478_half_map_1.map.gz emd_36478_half_map_1.map.gz emd_36478_half_map_2.map.gz emd_36478_half_map_2.map.gz | 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36478 http://ftp.pdbj.org/pub/emdb/structures/EMD-36478 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36478 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36478 | HTTPS FTP |
-Related structure data
| Related structure data |  8jpfMC  8jpbC  8jpcC  8jpdC  8jpeC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36478.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36478.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.824 Å | ||||||||||||||||||||
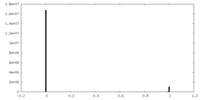
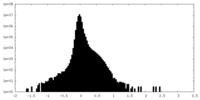
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_36478_msk_1.map emd_36478_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
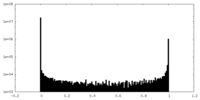
| Density Histograms |
-Half map: #1
| File | emd_36478_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
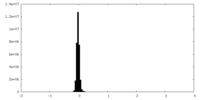
| Density Histograms |
-Half map: #2
| File | emd_36478_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
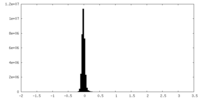
| Density Histograms |
- Sample components
Sample components
-Entire : Focused refiment structure of NTSR1 in NTSR1-GRK2-Galpha(q) complexes
| Entire | Name: Focused refiment structure of NTSR1 in NTSR1-GRK2-Galpha(q) complexes |
|---|---|
| Components |
|
-Supramolecule #1: Focused refiment structure of NTSR1 in NTSR1-GRK2-Galpha(q) complexes
| Supramolecule | Name: Focused refiment structure of NTSR1 in NTSR1-GRK2-Galpha(q) complexes type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: NTS
| Macromolecule | Name: NTS / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 819.007 Da |
| Sequence | String: RRPYIL |
-Macromolecule #2: Neurotensin receptor type 1
| Macromolecule | Name: Neurotensin receptor type 1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 46.307594 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MRLNSSAPGT PGTPAADPFQ RAQAGLEEAL LAPGFGNASG NASERVLAAP SSELDVNTDI YSKVLVTAVY LALFVVGTVG NTVTAFTLA RKKSLQSLQS TVHYHLGSLA LSDLLTLLLA MPVELYNFIW VHHPWAFGDA GCRGYYFLRD ACTYATALNV A SLSVERYL ...String: MRLNSSAPGT PGTPAADPFQ RAQAGLEEAL LAPGFGNASG NASERVLAAP SSELDVNTDI YSKVLVTAVY LALFVVGTVG NTVTAFTLA RKKSLQSLQS TVHYHLGSLA LSDLLTLLLA MPVELYNFIW VHHPWAFGDA GCRGYYFLRD ACTYATALNV A SLSVERYL AICHPFKAKT LMSRSRTKKF ISAIWLASAL LAVPMLFTMG EQNRSADGQH AGGLVCTPTI HTATVKVVIQ VN TFMSFIF PMVVISVLNT IIANKLTVMV RQAAEQGQVC TVGGEHSTFS MAIEPGRVQA LRHGVRVLRA VVIAFVVCWL PYH VRRLMF CYISDEQWTP FLYDFYHYFY MVTNALFYVS STINPILYNL VSANFRHIFL ATLACLCPVW RRRRKRPAFS RKAD SVSSN HTLSSNATRE TLY UniProtKB: Neurotensin receptor type 1 |
-Macromolecule #3: 2-[{2-(1-fluorocyclopropyl)-4-[4-(2-methoxyphenyl)piperidin-1-yl]...
| Macromolecule | Name: 2-[{2-(1-fluorocyclopropyl)-4-[4-(2-methoxyphenyl)piperidin-1-yl]quinazolin-6-yl}(methyl)amino]ethan-1-ol type: ligand / ID: 3 / Number of copies: 1 / Formula: SRW |
|---|---|
| Molecular weight | Theoretical: 450.548 Da |
| Chemical component information |  ChemComp-SRW: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DARK FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.02 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 474232 |
 Movie
Movie Controller
Controller


















 Z
Z Y
Y X
X