+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of inactive FZD1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Frizzled / class-F / FZD1 / Frizzled / class-F / FZD1 /  Complex / Complex /  MEMBRANE PROTEIN MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information : / muscular septum morphogenesis / regulation of mesenchymal stem cell differentiation / : / muscular septum morphogenesis / regulation of mesenchymal stem cell differentiation /  autocrine signaling / planar cell polarity pathway involved in neural tube closure / astrocyte-dopaminergic neuron signaling / canonical Wnt signaling pathway involved in mesenchymal stem cell differentiation / hard palate development / autocrine signaling / planar cell polarity pathway involved in neural tube closure / astrocyte-dopaminergic neuron signaling / canonical Wnt signaling pathway involved in mesenchymal stem cell differentiation / hard palate development /  Wnt receptor activity / membranous septum morphogenesis ... Wnt receptor activity / membranous septum morphogenesis ... : / muscular septum morphogenesis / regulation of mesenchymal stem cell differentiation / : / muscular septum morphogenesis / regulation of mesenchymal stem cell differentiation /  autocrine signaling / planar cell polarity pathway involved in neural tube closure / astrocyte-dopaminergic neuron signaling / canonical Wnt signaling pathway involved in mesenchymal stem cell differentiation / hard palate development / autocrine signaling / planar cell polarity pathway involved in neural tube closure / astrocyte-dopaminergic neuron signaling / canonical Wnt signaling pathway involved in mesenchymal stem cell differentiation / hard palate development /  Wnt receptor activity / membranous septum morphogenesis / non-canonical Wnt signaling pathway / presynapse assembly / Wnt-protein binding / endothelial cell differentiation / midbrain dopaminergic neuron differentiation / negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway / Wnt receptor activity / membranous septum morphogenesis / non-canonical Wnt signaling pathway / presynapse assembly / Wnt-protein binding / endothelial cell differentiation / midbrain dopaminergic neuron differentiation / negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway /  frizzled binding / PCP/CE pathway / Wnt signalosome / Class B/2 (Secretin family receptors) / Disassembly of the destruction complex and recruitment of AXIN to the membrane / outflow tract morphogenesis / regulation of presynapse assembly / negative regulation of BMP signaling pathway / canonical Wnt signaling pathway / positive regulation of osteoblast differentiation / Asymmetric localization of PCP proteins / TCF dependent signaling in response to WNT / frizzled binding / PCP/CE pathway / Wnt signalosome / Class B/2 (Secretin family receptors) / Disassembly of the destruction complex and recruitment of AXIN to the membrane / outflow tract morphogenesis / regulation of presynapse assembly / negative regulation of BMP signaling pathway / canonical Wnt signaling pathway / positive regulation of osteoblast differentiation / Asymmetric localization of PCP proteins / TCF dependent signaling in response to WNT /  PDZ domain binding / G protein-coupled receptor activity / negative regulation of canonical Wnt signaling pathway / neuron differentiation / positive regulation of neuron projection development / positive regulation of DNA-binding transcription factor activity / cell-cell signaling / PDZ domain binding / G protein-coupled receptor activity / negative regulation of canonical Wnt signaling pathway / neuron differentiation / positive regulation of neuron projection development / positive regulation of DNA-binding transcription factor activity / cell-cell signaling /  electron transfer activity / electron transfer activity /  periplasmic space / response to xenobiotic stimulus / positive regulation of protein phosphorylation / iron ion binding / periplasmic space / response to xenobiotic stimulus / positive regulation of protein phosphorylation / iron ion binding /  signaling receptor binding / signaling receptor binding /  focal adhesion / negative regulation of DNA-templated transcription / focal adhesion / negative regulation of DNA-templated transcription /  heme binding / positive regulation of DNA-templated transcription / heme binding / positive regulation of DNA-templated transcription /  cell surface / cell surface /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /   Mus (mice) / Mus (mice) /   Vicugna pacos (alpaca) Vicugna pacos (alpaca) | |||||||||
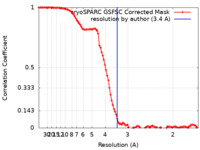
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.4 Å cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Lin X / Xu F | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2024 Journal: Cell Discov / Year: 2024Title: A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways. Authors: Zhibin Zhang / Xi Lin / Ling Wei / Yiran Wu / Lu Xu / Lijie Wu / Xiaohu Wei / Suwen Zhao / Xiangjia Zhu / Fei Xu /  Abstract: The ten Frizzled receptors (FZDs) are essential in Wnt signaling and play important roles in embryonic development and tumorigenesis. Among these, FZD6 is closely associated with lens development. ...The ten Frizzled receptors (FZDs) are essential in Wnt signaling and play important roles in embryonic development and tumorigenesis. Among these, FZD6 is closely associated with lens development. Understanding FZD activation mechanism is key to unlock these emerging targets. Here we present the cryo-EM structures of FZD6 and FZD3 which are known to relay non-canonical planar cell polarity (PCP) signaling pathways as well as FZD1 in their G protein-coupled states and in the apo inactive states, respectively. Comparison of the three inactive/active pairs unveiled a shared activation framework among all ten FZDs. Mutagenesis along with imaging and functional analysis on the human lens epithelial tissues suggested potential crosstalk between the G-protein coupling of FZD6 and the PCP signaling pathways. Together, this study provides an integrated understanding of FZD structure and function, and lays the foundation for developing therapeutic modulators to activate or inhibit FZD signaling for a range of disorders including cancers and cataracts. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36112.map.gz emd_36112.map.gz | 80.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36112-v30.xml emd-36112-v30.xml emd-36112.xml emd-36112.xml | 19.1 KB 19.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36112_fsc.xml emd_36112_fsc.xml | 10.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_36112.png emd_36112.png | 40.4 KB | ||
| Filedesc metadata |  emd-36112.cif.gz emd-36112.cif.gz | 6.2 KB | ||
| Others |  emd_36112_additional_1.map.gz emd_36112_additional_1.map.gz emd_36112_half_map_1.map.gz emd_36112_half_map_1.map.gz emd_36112_half_map_2.map.gz emd_36112_half_map_2.map.gz | 85.8 MB 84.7 MB 84.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36112 http://ftp.pdbj.org/pub/emdb/structures/EMD-36112 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36112 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36112 | HTTPS FTP |
-Related structure data
| Related structure data |  8j9oMC  8j9nC  8jh7C  8jhbC  8jhcC  8jhiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36112.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36112.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_36112_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_36112_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_36112_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Frizzled-1 with ICL3-BRIL complex with anti-BRIL Fab
| Entire | Name: Frizzled-1 with ICL3-BRIL complex with anti-BRIL Fab |
|---|---|
| Components |
|
-Supramolecule #1: Frizzled-1 with ICL3-BRIL complex with anti-BRIL Fab
| Supramolecule | Name: Frizzled-1 with ICL3-BRIL complex with anti-BRIL Fab / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Frizzled-1,Soluble cytochrome b562
| Macromolecule | Name: Frizzled-1,Soluble cytochrome b562 / type: protein_or_peptide / ID: 1 / Details: inactive FZD1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 75.034164 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: GVRAQAAGQG PGQGPGPGQQ PPPPPQQQQS GQQYNGERGI SVPDHGYCQP ISIPLCTDIA YNQTIMPNLL GHTNQEDAGL EVHQFYPLV KVQCSAELKF FLCSMYAPVC TVLEQALPPC RSLCERARQG CEALMNKFGF QWPDTLKCEK FPVHGAGELC V GQNTSDKG ...String: GVRAQAAGQG PGQGPGPGQQ PPPPPQQQQS GQQYNGERGI SVPDHGYCQP ISIPLCTDIA YNQTIMPNLL GHTNQEDAGL EVHQFYPLV KVQCSAELKF FLCSMYAPVC TVLEQALPPC RSLCERARQG CEALMNKFGF QWPDTLKCEK FPVHGAGELC V GQNTSDKG TPTPSLLPEF WTSNPQHGGG GHRGGFPGGA GASERGKFSC PRALKVPSYL NYHFLGEKDC GAPCEPTKVY GL MYFGPEE LRFSRTWIGI WSVLCCASTL FTVLTYLVDM RRFSYPERPI IFLSGCYTAV AVAYIAGFLL EDRVVCNDKF AED GARTVA QGTKKEGCTI LFMMLYFFSM ASSIWWVILS LTWFLAAGMK WGHEAIEANS QYFHLAAWAV PAIKTITILA LGQV DGDVL SGVCFVGLNN VDALRGFVLA PLFVYLFIGT SFLLAGFVSL FIRARRQLAD LEDNWETLND NLKVIEKADN AAQVK DALT KMRAAALDAQ KATPPKLEDK SPDSPEMKDF RHGFDILVGQ IDDALKLANE GKVKEAQAAA EQLKTTRNAY IQKYLE RAR STLKEKLEKL MVRIGVFSVL YTVPATIVIA CYFYEQAFRD QWERSWVAQS CKSYAIPCPH LQAGGGAPPH PPMSPDF TV FMIKYLMTLI VGITSGFWIW SGKTLNSWRK FYTRL UniProtKB:  Frizzled-1, Soluble cytochrome b562, Frizzled-1, Soluble cytochrome b562,  Frizzled-1 Frizzled-1 |
-Macromolecule #2: anti-BRIL Fab Heavy Chain
| Macromolecule | Name: anti-BRIL Fab Heavy Chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus (mice) Mus (mice) |
| Molecular weight | Theoretical: 28.03542 KDa |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MVSAIVLYVL LAAAAHSAFA EISEVQLVES GGGLVQPGGS LRLSCAASGF NVVDFSLHWV RQAPGKGLEW VAYISSSSGS TSYADSVKG RFTISADTSK NTAYLQMNSL RAEDTAVYYC ARWGYWPGEP WWKAFDYWGQ GTLVTVSSAS TKGPSVFPLA P SSKSTSGG ...String: MVSAIVLYVL LAAAAHSAFA EISEVQLVES GGGLVQPGGS LRLSCAASGF NVVDFSLHWV RQAPGKGLEW VAYISSSSGS TSYADSVKG RFTISADTSK NTAYLQMNSL RAEDTAVYYC ARWGYWPGEP WWKAFDYWGQ GTLVTVSSAS TKGPSVFPLA P SSKSTSGG TAALGCLVKD YFPEPVTVSW NSGALTSGVH TFPAVLQSSG LYSLSSVVTV PSSSLGTQTY ICNVNHKPSN TK VDKKVEP KSLEVLFQGP HHHHHH |
-Macromolecule #3: anti-Fab nanobody
| Macromolecule | Name: anti-Fab nanobody / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Vicugna pacos (alpaca) Vicugna pacos (alpaca) |
| Molecular weight | Theoretical: 13.390644 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: GSQVQLQESG GGLVQPGGSL RLSCAASGRT ISRYAMSWFR QAPGKEREFV AVARRSGDGA FYADSVQGRF TVSRDDAKNT VYLQMNSLK PEDTAVYYCA IDSDTFYSGS YDYWGQGTQV TVSS |
-Macromolecule #4: anti-BRIL Fab Light Chain
| Macromolecule | Name: anti-BRIL Fab Light Chain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus (mice) Mus (mice) |
| Molecular weight | Theoretical: 24.557408 KDa |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MVVYISYIYA SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSL QPEDFATYYC QQYLYYSLVT FGQGTKVEIK RTVAAPSVFI FPPSDSQLKS GTASVVCLLN NFYPREAKVQ W KVDNALQS ...String: MVVYISYIYA SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSL QPEDFATYYC QQYLYYSLVT FGQGTKVEIK RTVAAPSVFI FPPSDSQLKS GTASVVCLLN NFYPREAKVQ W KVDNALQS GNSQESVTEQ DSKDSTYSLS STLTLSKADY EKHKVYACEV THQGLSSPVT KSFNRG |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source: OTHER |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.7000000000000001 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z
Z Y
Y X
X