+ Open data
Open data
- Basic information
Basic information
| Entry | 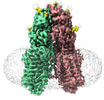 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | transport T2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transport T2 /  TRANSPORT PROTEIN TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnucleic acid transmembrane transporter activity / AP-1 adaptor complex binding / RNA transmembrane transporter activity / RNA transport / AP-2 adaptor complex binding / type B pancreatic cell development / regulation of insulin secretion involved in cellular response to glucose stimulus / type B pancreatic cell proliferation / RNA catabolic process / response to glucose ...nucleic acid transmembrane transporter activity / AP-1 adaptor complex binding / RNA transmembrane transporter activity / RNA transport / AP-2 adaptor complex binding / type B pancreatic cell development / regulation of insulin secretion involved in cellular response to glucose stimulus / type B pancreatic cell proliferation / RNA catabolic process / response to glucose /  bioluminescence / generation of precursor metabolites and energy / cell morphogenesis / bioluminescence / generation of precursor metabolites and energy / cell morphogenesis /  glucose homeostasis / glucose homeostasis /  double-stranded RNA binding / double-stranded RNA binding /  lysosome / lysosomal membrane / lysosome / lysosomal membrane /  DNA binding / DNA binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.25 Å cryo EM / Resolution: 3.25 Å | |||||||||
 Authors Authors | Jiang DH / Zhang JT | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structural insights into double-stranded RNA recognition and transport by SID-1. Authors: Jiangtao Zhang / Chunhua Zhan / Junping Fan / Dian Wu / Ruixue Zhang / Di Wu / Xinyao Chen / Ying Lu / Ming Li / Min Lin / Jianke Gong / Daohua Jiang /  Abstract: RNA uptake by cells is critical for RNA-mediated gene interference (RNAi) and RNA-based therapeutics. In Caenorhabditis elegans, RNAi is systemic as a result of SID-1-mediated double-stranded RNA ...RNA uptake by cells is critical for RNA-mediated gene interference (RNAi) and RNA-based therapeutics. In Caenorhabditis elegans, RNAi is systemic as a result of SID-1-mediated double-stranded RNA (dsRNA) across cells. Despite the functional importance, the underlying mechanisms of dsRNA internalization by SID-1 remain elusive. Here we describe cryogenic electron microscopy structures of SID-1, SID-1-dsRNA complex and human SID-1 homologs SIDT1 and SIDT2, elucidating the structural basis of dsRNA recognition and import by SID-1. The homodimeric SID-1 homologs share conserved architecture, but only SID-1 possesses the molecular determinants within its extracellular domains for distinguishing dsRNA from single-stranded RNA and DNA. We show that the removal of the long intracellular loop between transmembrane helix 1 and 2 attenuates dsRNA uptake and systemic RNAi in vivo, suggesting a possible endocytic mechanism of SID-1-mediated dsRNA internalization. Our study provides mechanistic insights into dsRNA internalization by SID-1, which may facilitate the development of dsRNA applications based on SID-1. | |||||||||
| History |
|
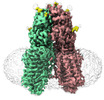
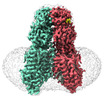
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36009.map.gz emd_36009.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36009-v30.xml emd-36009-v30.xml emd-36009.xml emd-36009.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36009.png emd_36009.png | 171.1 KB | ||
| Filedesc metadata |  emd-36009.cif.gz emd-36009.cif.gz | 6 KB | ||
| Others |  emd_36009_half_map_1.map.gz emd_36009_half_map_1.map.gz emd_36009_half_map_2.map.gz emd_36009_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36009 http://ftp.pdbj.org/pub/emdb/structures/EMD-36009 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36009 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36009 | HTTPS FTP |
-Related structure data
| Related structure data |  8j6oMC  8hipC  8hkeC  8j6mC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36009.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36009.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_36009_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
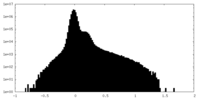
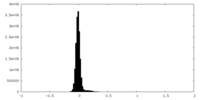
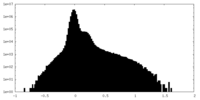
| Density Histograms |
-Half map: #2
| File | emd_36009_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : T2
| Entire | Name: T2 |
|---|---|
| Components |
|
-Supramolecule #1: T2
| Supramolecule | Name: T2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Green fluorescent protein (Fragment),SID1 transmembrane family me...
| Macromolecule | Name: Green fluorescent protein (Fragment),SID1 transmembrane family member 2 type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 124.883797 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MYRMQLLSCI ALSLALVTNS WSHPQFEKGG GSGGGSGGSA WSHPQFEKSK GEELFTGVVP ILVELDGDVN GHKFSVRGEG EGDATNGKL TLKFICTTGK LPVPWPTLVT TLTYGVQCFS RYPDHMKRHD FFKSAMPEGY VQERTISFKD DGTYKTRAEV K FEGDTLVN ...String: MYRMQLLSCI ALSLALVTNS WSHPQFEKGG GSGGGSGGSA WSHPQFEKSK GEELFTGVVP ILVELDGDVN GHKFSVRGEG EGDATNGKL TLKFICTTGK LPVPWPTLVT TLTYGVQCFS RYPDHMKRHD FFKSAMPEGY VQERTISFKD DGTYKTRAEV K FEGDTLVN RIELKGIDFK EDGNILGHKL EYNFNSHNVY ITADKQKNGI KANFKIRHNV EDGSVQLADH YQQNTPIGDG PV LLPDNHY LSTQSVLSKD PNEKRDHMVL LEFVTAAGIT HGMDELEVLF QGPHLGVLGP KNVSQKDAEF ERTYVDEVNS ELV NIYTFN HTVTRNRTEG VRVSVNVLNK QKGAPLLFVV RQKEAVVSFQ VPLILRGMFQ RKYLYQKVER TLCQPPTKNE SEIQ FFYVD VSTLSPVNTT YQLRVSRMDD FVLRTGEQFS FNTTAAQPQY FKYEFPEGVD SVIVKVTSNK AFPCSVISIQ DVLCP VYDL DNNVAFIGMY QTMTKKAAIT VQRKDFPSNS FYVVVVVKTE DQACGGSLPF YPFAEDEPVD QGHRQKTLSV LVSQAV TSE AYVSGMLFCL GIFLSFYLLT VLLACWENWR QKKKTLLVAI DRACPESGHP RVLADSFPGS SPYEGYNYGS FENVSGS TD GLVDSAGTGD LSYGYQGRSF EPVGTRPRVD SMSSVEEDDY DTLTDIDSDK NVIRTKQYLY VADLARKDKR VLRKKYQI Y FWNIATIAVF YALPVVQLVI TYQTVVNVTG NQDICYYNFL CAHPLGNLSA FNNILSNLGY ILLGLLFLLI ILQREINHN RALLRNDLCA LECGIPKHFG LFYAMGTALM MEGLLSACYH VCPNYTNFQF DTSFMYMIAG LCMLKLYQKR HPDINASAYS AYACLAIVI FFSVLGVVFG KGNTAFWIVF SIIHIIATLL LSTQLYYMGR WKLDSGIFRR ILHVLYTDCI RQCSGPLYVD R MVLLVMGN VINWSLAAYG LIMRPNDFAS YLLAIGICNL LLYFAFYIIM KLRSGERIKL IPLLCIVCTS VVWGFALFFF FQ GLSTWQK TPAESREHNR DCILLDFFDD HDIWHFLSSI AMFGSFLVLL TLDDDLDTVQ RDKIYVF UniProtKB:  Green fluorescent protein, SID1 transmembrane family member 2 Green fluorescent protein, SID1 transmembrane family member 2 |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 4 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm Bright-field microscopy / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.25 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 104974 |
 Movie
Movie Controller
Controller







 Z
Z Y
Y X
X