[English] 日本語
 Yorodumi
Yorodumi- EMDB-35426: Cryo-EM structure of SARS-CoV-2 Omicron BA.4/5 spike protein in c... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of SARS-CoV-2 Omicron BA.4/5 spike protein in complex with white-tailed deer ACE2 | |||||||||
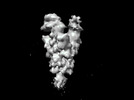
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  complex / complex /  VIRAL PROTEIN / VIRAL PROTEIN /  VIRAL PROTEIN-HYDROLASE complex VIRAL PROTEIN-HYDROLASE complex | |||||||||
| Function / homology |  Function and homology information Function and homology information Hydrolases; Acting on peptide bonds (peptidases) / peptidyl-dipeptidase activity / Hydrolases; Acting on peptide bonds (peptidases) / peptidyl-dipeptidase activity /  carboxypeptidase activity / carboxypeptidase activity /  metallopeptidase activity / Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space ... metallopeptidase activity / Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space ... Hydrolases; Acting on peptide bonds (peptidases) / peptidyl-dipeptidase activity / Hydrolases; Acting on peptide bonds (peptidases) / peptidyl-dipeptidase activity /  carboxypeptidase activity / carboxypeptidase activity /  metallopeptidase activity / Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / receptor-mediated endocytosis of virus by host cell / Attachment and Entry / metallopeptidase activity / Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / receptor-mediated endocytosis of virus by host cell / Attachment and Entry /  membrane fusion / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / membrane fusion / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell /  receptor ligand activity / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / receptor ligand activity / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / viral envelope / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane /  proteolysis / proteolysis /  membrane / identical protein binding / membrane / identical protein binding /  metal ion binding / metal ion binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Severe acute respiratory syndrome coronavirus 2 / Severe acute respiratory syndrome coronavirus 2 /   Odocoileus virginianus (white-tailed deer) Odocoileus virginianus (white-tailed deer) | |||||||||
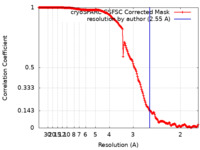
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.55 Å cryo EM / Resolution: 2.55 Å | |||||||||
 Authors Authors | Han P / Meng YM / Qi JX | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: J Virol / Year: 2023 Journal: J Virol / Year: 2023Title: Structural basis of white-tailed deer, , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity. Authors: Pu Han / Yumin Meng / Di Zhang / Zepeng Xu / Zhiyuan Li / Xiaoqian Pan / Zhennan Zhao / Linjie Li / Lingfeng Tang / Jianxun Qi / Kefang Liu / George F Gao /  Abstract: SARS-CoV-2 has been expanding its host range, among which the white-tailed deer (WTD), became the first wildlife species infected on a large scale and might serve as a host reservoir for variants of ...SARS-CoV-2 has been expanding its host range, among which the white-tailed deer (WTD), became the first wildlife species infected on a large scale and might serve as a host reservoir for variants of concern (VOCs) in case no longer circulating in humans. In this study, we comprehensively assessed the binding of the WTD angiotensin-converting enzyme 2 (ACE2) receptor to the spike (S) receptor-binding domains (RBDs) from the SARS-CoV-2 prototype (PT) strain and multiple variants. We found that WTD ACE2 could be broadly recognized by all of the tested RBDs. We further determined the complex structures of WTD ACE2 with PT, Omicron BA.1, and BA.4/5 S trimer. Detailed structural comparison revealed the important roles of RBD residues on 486, 498, and 501 sites for WTD ACE2 binding. This study deepens our understanding of the interspecies transmission mechanisms of SARS-CoV-2 and further addresses the importance of constant monitoring on SARS-CoV-2 infections in wild animals. IMPORTANCE Even if we manage to eliminate the virus among humans, it will still circulate among wildlife and continuously be transmitted back to humans. A recent study indicated that WTD may serve as reservoir for nearly extinct SARS-CoV-2 strains. Therefore, it is critical to evaluate the binding abilities of SARS-CoV-2 variants to the WTD ACE2 receptor and elucidate the molecular mechanisms of binding of the RBDs to assess the risk of spillback events. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35426.map.gz emd_35426.map.gz | 268.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35426-v30.xml emd-35426-v30.xml emd-35426.xml emd-35426.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35426_fsc.xml emd_35426_fsc.xml | 17 KB | Display |  FSC data file FSC data file |
| Images |  emd_35426.png emd_35426.png | 33.7 KB | ||
| Filedesc metadata |  emd-35426.cif.gz emd-35426.cif.gz | 6.8 KB | ||
| Others |  emd_35426_half_map_1.map.gz emd_35426_half_map_1.map.gz emd_35426_half_map_2.map.gz emd_35426_half_map_2.map.gz | 498.5 MB 498.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35426 http://ftp.pdbj.org/pub/emdb/structures/EMD-35426 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35426 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35426 | HTTPS FTP |
-Related structure data
| Related structure data |  8ifyMC  8hfxC  8hfyC  8hfzC  8hg0C  8ifzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35426.map.gz / Format: CCP4 / Size: 536.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35426.map.gz / Format: CCP4 / Size: 536.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.88 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_35426_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_35426_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of SARS-CoV-2 Omicron BA.4/5 spike protein in c...
| Entire | Name: Cryo-EM structure of SARS-CoV-2 Omicron BA.4/5 spike protein in complex with white-tailed deer ACE2 |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of SARS-CoV-2 Omicron BA.4/5 spike protein in c...
| Supramolecule | Name: Cryo-EM structure of SARS-CoV-2 Omicron BA.4/5 spike protein in complex with white-tailed deer ACE2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Severe acute respiratory syndrome coronavirus 2 Severe acute respiratory syndrome coronavirus 2 |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Severe acute respiratory syndrome coronavirus 2 Severe acute respiratory syndrome coronavirus 2 |
| Molecular weight | Theoretical: 125.148039 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QCVNLITRTQ SYTNSFTRGV YYPDKVFRSS VLHSTQDLFL PFFSNVTWFH AISGTNGTKR FDNPVLPFND GVYFASTEKS NIIRGWIFG TTLDSKTQSL LIVNNATNVV IKVCEFQFCN DPFLDVYYHK NNKSWMESEF RVYSSANNCT FEYVSQPFLM D LEGKQGNF ...String: QCVNLITRTQ SYTNSFTRGV YYPDKVFRSS VLHSTQDLFL PFFSNVTWFH AISGTNGTKR FDNPVLPFND GVYFASTEKS NIIRGWIFG TTLDSKTQSL LIVNNATNVV IKVCEFQFCN DPFLDVYYHK NNKSWMESEF RVYSSANNCT FEYVSQPFLM D LEGKQGNF KNLREFVFKN IDGYFKIYSK HTPINLGRDL PQGFSALEPL VDLPIGINIT RFQTLLALHR SYLTPGDSSS GW TAGAAAY YVGYLQPRTF LLKYNENGTI TDAVDCALDP LSETKCTLKS FTVEKGIYQT SNFRVQPTES IVRFPNITNL CPF DEVFNA TRFASVYAWN RKRISNCVAD YSVLYNFAPF FAFKCYGVSP TKLNDLCFTN VYADSFVIRG NEVSQIAPGQ TGNI ADYNY KLPDDFTGCV IAWNSNKLDS KVGGNYNYRY RLFRKSNLKP FERDISTEIY QAGNKPCNGV AGVNCYFPLQ SYGFR PTYG VGHQPYRVVV LSFELLHAPA TVCGPKKSTN LVKNKCVNFN FNGLTGTGVL TESNKKFLPF QQFGRDIADT TDAVRD PQT LEILDITPCS FGGVSVITPG TNTSNQVAVL YQGVNCTEVP VAIHADQLTP TWRVYSTGSN VFQTRAGCLI GAEYVNN SY ECDIPIGAGI CASYQTQTKS HRRARSVASQ SIIAYTMSLG AENSVAYSNN SIAIPTNFTI SVTTEILPVS MTKTSVDC T MYICGDSTEC SNLLLQYGSF CTQLKRALTG IAVEQDKNTQ EVFAQVKQIY KTPPIKYFGG FNFSQILPDP SKPSKRSPI EDLLFNKVTL ADAGFIKQYG DCLGDIAARD LICAQKFNGL TVLPPLLTDE MIAQYTSALL AGTITSGWTF GAGPALQIPF PMQMAYRFN GIGVTQNVLY ENQKLIANQF NSAIGKIQDS LSSTPSALGK LQDVVNHNAQ ALNTLVKQLS SKFGAISSVL N DILSRLDP PEAEVQIDRL ITGRLQSLQT YVTQQLIRAA EIRASANLAA TKMSECVLGQ SKRVDFCGKG YHLMSFPQSA PH GVVFLHV TYVPAQEKNF TTAPAICHDG KAHFPREGVF VSNGTHWFVT QRNFYEPQII TTDNTFVSGN CDVVIGIVNN TVY DPLQPE L UniProtKB:  Spike glycoprotein Spike glycoprotein |
-Macromolecule #2: Angiotensin-converting enzyme
| Macromolecule | Name: Angiotensin-converting enzyme / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Odocoileus virginianus (white-tailed deer) Odocoileus virginianus (white-tailed deer) |
| Molecular weight | Theoretical: 76.530477 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: STTEEQAKTF LEKFNHEAED LSYQSSLASW NYNTNITDEN VQKMNEARAK WSAFYEEQSR MAKTYSLEEI QNLTLKRQLK ALQQSGTSV LSAEKSKRLN TILNTMSTIY STGKVLDPNT QECLALEPGL DDIMENSRDY NRRLWAWEGW RAEVGKQLRP L YEEYVVLE ...String: STTEEQAKTF LEKFNHEAED LSYQSSLASW NYNTNITDEN VQKMNEARAK WSAFYEEQSR MAKTYSLEEI QNLTLKRQLK ALQQSGTSV LSAEKSKRLN TILNTMSTIY STGKVLDPNT QECLALEPGL DDIMENSRDY NRRLWAWEGW RAEVGKQLRP L YEEYVVLE NEMARANNYE DYGDYWRGDY EVTEAGDYDY SRDQLMKDVE NTFAEIKPLY EQLHAYVRAK LMDTYPSYIS PT GCLPAHL LGDMWGRFWT NLYSLTVPFK HKPSIDVTEK MKNQSWDAER IFKEAEKFFV SISLPHMTQG FWDNSMLTEP GDG RKVVCH PTAWDLGKGD FRIKMCTKVT MDDFLTAHHE MGHIQYDMAY AAQPYLLRDG ANEGFHEAVG EIMSLSAATP HYLK ALGLL EPDFYEDNET EINFLLKQAL TIVGTLPFTY MLEKWRWMVF KGEIPKEQWM EKWWEMKREI VGVVEPLPHD ETYCD PACL FHVAEDYSFI RYYTRTIYQF QFHEALCKTA NHEGALFKCD ISNSTEAGQR LLQMLSLGKS EPWTLALESI VGIKTM DVK PLLNYFEPLF TWLKEQNRNS FVGWSTEWTP YSDQSIKVRI SLKSALGKNA DANCPFVWCV PPVSHLVAIV IRSAVTV SQ CCVQATLVLL NPGPKVPEE UniProtKB:  Angiotensin-converting enzyme Angiotensin-converting enzyme |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 25 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #5: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z
Z Y
Y X
X