[English] 日本語
 Yorodumi
Yorodumi- EMDB-35402: Conformational Dynamics of the D53-D3-D14 Complex in Strigolacton... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Conformational Dynamics of the D53-D3-D14 Complex in Strigolactone Signaling | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | D14 D3 D53 SL /  PLANT PROTEIN PLANT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationbud dilation /  regulation of shoot system morphogenesis / regulation of shoot system morphogenesis /  shoot system morphogenesis / regulation of meristem structural organization / negative regulation of seed germination / positive regulation of response to water deprivation / strigolactone biosynthetic process / cuticle development / auxin polar transport / secondary shoot formation ...bud dilation / shoot system morphogenesis / regulation of meristem structural organization / negative regulation of seed germination / positive regulation of response to water deprivation / strigolactone biosynthetic process / cuticle development / auxin polar transport / secondary shoot formation ...bud dilation /  regulation of shoot system morphogenesis / regulation of shoot system morphogenesis /  shoot system morphogenesis / regulation of meristem structural organization / negative regulation of seed germination / positive regulation of response to water deprivation / strigolactone biosynthetic process / cuticle development / auxin polar transport / secondary shoot formation / jasmonic acid mediated signaling pathway / response to water deprivation / SCF-dependent proteasomal ubiquitin-dependent protein catabolic process / shoot system morphogenesis / regulation of meristem structural organization / negative regulation of seed germination / positive regulation of response to water deprivation / strigolactone biosynthetic process / cuticle development / auxin polar transport / secondary shoot formation / jasmonic acid mediated signaling pathway / response to water deprivation / SCF-dependent proteasomal ubiquitin-dependent protein catabolic process /  SCF ubiquitin ligase complex / cullin family protein binding / response to light stimulus / SCF ubiquitin ligase complex / cullin family protein binding / response to light stimulus /  Hydrolases; Acting on ester bonds / protein ubiquitination / Hydrolases; Acting on ester bonds / protein ubiquitination /  hydrolase activity / hydrolase activity /  nucleus / nucleus /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Oryza sativa (Asian cultivated rice) / Oryza sativa (Asian cultivated rice) /   Oryza sativa subsp. japonica (Japanese rice) Oryza sativa subsp. japonica (Japanese rice) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 7.09 Å cryo EM / Resolution: 7.09 Å | |||||||||
 Authors Authors | Liu SM / Wang J | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Plant Cell Physiol / Year: 2023 Journal: Plant Cell Physiol / Year: 2023Title: Conformational Dynamics of the D53-D3-D14 Complex in Strigolactone Signaling. Authors: Simiao Liu / Jia Wang / Bin Song / Xinqi Gong / Huihui Liu / Qingliang Hu / Junhui Zhang / Qianqian Li / Jie Zheng / Hongwei Wang / H Eric Xu / Jiayang Li / Bing Wang /  Abstract: Strigolactones (SLs) play fundamental roles in regulating plant architecture, which is a major factor determining crop yield. The perception and signal transduction of SLs require the formation of a ...Strigolactones (SLs) play fundamental roles in regulating plant architecture, which is a major factor determining crop yield. The perception and signal transduction of SLs require the formation of a complex containing the receptor DWARF14 (D14), an F-box protein D3 and a transcriptional regulator D53 in an SL-dependent manner. Structural and biochemical analyses of D14 and its orthologs DAD2 and AtD14, D3 and the complexes of ASK1-D3-AtD14 and D3CTH-D14 have made great contributions to understanding the mechanisms of SL perception. However, structural analyses of D53 and the D53-D3-D14 holo-complex are challenging, and the biochemical mechanism underlying the complex assembly remains poorly understood. Here, we found that apo-D53 was rather flexible and reconstituted the holo-complex containing D53, S-phase kinase-associated protein 1 (SKP1), D3 and D14 with rac-GR24. The cryo-electron microscopy (cryo-EM) structure of SKP1-D3-D14 in the presence of D53 was analyzed and superimposed on the crystal structure of ASK1-D3-AtD14 without D53. No large conformational rearrangement was observed, but a 9Å rotation appeared between D14 and AtD14. Using hydrogen-deuterium exchange monitored by mass spectrometry, we analyzed dynamic motifs of D14, D3 and D53 in the D53-SKP1-D3-D14 complex assembly process and further identified two potential interfaces in D53 that are located in the N and D2 domains, respectively. Together, our results uncovered the dynamic conformational changes and built a model of the holo-complex D53-SKP1-D3-D14, offering valuable information for the biochemical and genetic mechanisms of SL perception and signal transduction. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35402.map.gz emd_35402.map.gz | 1.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35402-v30.xml emd-35402-v30.xml emd-35402.xml emd-35402.xml | 16.4 KB 16.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35402.png emd_35402.png | 69 KB | ||
| Others |  emd_35402_half_map_1.map.gz emd_35402_half_map_1.map.gz emd_35402_half_map_2.map.gz emd_35402_half_map_2.map.gz | 2.6 MB 2.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35402 http://ftp.pdbj.org/pub/emdb/structures/EMD-35402 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35402 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35402 | HTTPS FTP |
-Related structure data
| Related structure data |  8if6MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35402.map.gz / Format: CCP4 / Size: 2.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35402.map.gz / Format: CCP4 / Size: 2.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 2.182 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_35402_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
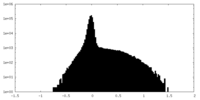
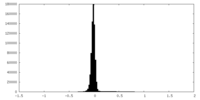
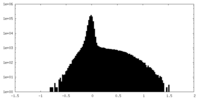
| Density Histograms |
-Half map: #2
| File | emd_35402_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Plant protein complex
| Entire | Name: Plant protein complex |
|---|---|
| Components |
|
-Supramolecule #1: Plant protein complex
| Supramolecule | Name: Plant protein complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Oryza sativa (Asian cultivated rice) Oryza sativa (Asian cultivated rice) |
-Macromolecule #1: F-box/LRR-repeat MAX2 homolog
| Macromolecule | Name: F-box/LRR-repeat MAX2 homolog / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Oryza sativa subsp. japonica (Japanese rice) Oryza sativa subsp. japonica (Japanese rice) |
| Molecular weight | Theoretical: 79.308617 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MAEEEEVEEG RSSSSAILDL PEPLLLHILS FLTDVRSRHR AALACGRMRA AERATRSELS LRGDPRSPGF LFLSHAFRFP ALEHLDLSL VSPWGHPLLS SVPPCGGGGG GAPSASSSSG MNVYHPEAIS EQNAFIAARL AGCFPAVTSL AVYCRDPTTL A NLTPHWQA ...String: MAEEEEVEEG RSSSSAILDL PEPLLLHILS FLTDVRSRHR AALACGRMRA AERATRSELS LRGDPRSPGF LFLSHAFRFP ALEHLDLSL VSPWGHPLLS SVPPCGGGGG GAPSASSSSG MNVYHPEAIS EQNAFIAARL AGCFPAVTSL AVYCRDPTTL A NLTPHWQA SLRRVKLVRW HQRPPTLPDG ADLEPLLETC AALRELDLSE FYCWTEDVVR ALTTHPSATA ALTHLDLGLA AA TDGFKSS ELGPIAASCP NLRKLVAPCL FNPRFSDCVG DDALLSLATS CPRLTVLRLS EPFEAAANIQ REEAAITVAG LVA FFAALP ALEDFTMDLQ HNVLEAAPAM EALARRCPRI KFLTLGSFQG LCKASWLHLD GVAVCGGLES LYMKNCQDLT DASL AAIGR GCRRLAKFGI HGCDLVTSAG IRRLAFTLRP TLKEVTVLHC RLLHTAECLT ALSPIRDRIE SLEINCVWNT TEQPC SVAN GTTTECDPED DELGEVYESA AKKCRYMEFD DLGSWEMLRS LSLWFSAGQL LSPLISAGLD SCPVLEEISI KVEGDC RTC PRPAPRTIFG LSDLAGFPVL AKMKLDLSEA VGYALTAPTG QMDLSLWERF YLHGIESLQT LYELDYWPPQ DKDVHHR SL TLPAVGLIQR CVGLRKLFIH GTTHEHFMTF FLSIPNLRDM QLREDYYPAP ENDLMFTEMR AESWLRFEVQ LNSRQIDD UniProtKB: F-box/LRR-repeat MAX2 homolog |
-Macromolecule #2: Strigolactone esterase D14
| Macromolecule | Name: Strigolactone esterase D14 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number:  Hydrolases; Acting on ester bonds Hydrolases; Acting on ester bonds |
|---|---|
| Source (natural) | Organism:   Oryza sativa subsp. japonica (Japanese rice) Oryza sativa subsp. japonica (Japanese rice) |
| Molecular weight | Theoretical: 33.545023 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MLRSTHPPPS SPSSSSSGGG GGGGSSASSS SEKTMVGGGG GGGGGSGSAA PSGAKLLQIL NVRVVGSGER VVVLSHGFGT DQSAWSRVL PYLTRDHRVV LYDLVCAGSV NPDHFDFRRY DNLDAYVDDL LAILDALRIP RCAFVGHSVS AMIGILASIR R PDLFAKLV ...String: MLRSTHPPPS SPSSSSSGGG GGGGSSASSS SEKTMVGGGG GGGGGSGSAA PSGAKLLQIL NVRVVGSGER VVVLSHGFGT DQSAWSRVL PYLTRDHRVV LYDLVCAGSV NPDHFDFRRY DNLDAYVDDL LAILDALRIP RCAFVGHSVS AMIGILASIR R PDLFAKLV LIGASPRFLN DSDYHGGFEL EEIQQVFDAM GANYSAWATG YAPLAVGADV PAAVQEFSRT LFNMRPDISL HV CQTVFKT DLRGVLGMVR APCVVVQTTR DVSVPASVAA YLKAHLGGRT TVEFLQTEGH LPHLSAPSLL AQVLRRALAR Y UniProtKB: Strigolactone esterase D14 |
-Macromolecule #3: SKP1-like protein 20
| Macromolecule | Name: SKP1-like protein 20 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Oryza sativa subsp. japonica (Japanese rice) Oryza sativa subsp. japonica (Japanese rice) |
| Molecular weight | Theoretical: 19.217693 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MAAEAETKAM ITLRSCEGQV FEVAEAVAME SQTIRHMIED KCADTGIPLP NVSAKILSKV IEYCSKHVEA RGGAAAAADG DAPAPAAVE ANKAVEDELK TFDAEFVKVD QSTLFDLILA ANYLNIKGLL DLTCQTVADM IKGKTPEEIR KTFNIKNDFT P EEEEEVRR ENQWAFE UniProtKB: SKP1-like protein 20 |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 0.7000000000000001 µm / Nominal defocus min: 0.5 µm Bright-field microscopy / Nominal defocus max: 0.7000000000000001 µm / Nominal defocus min: 0.5 µm |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 7.09 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 42499 |
 Movie
Movie Controller
Controller











 Z
Z Y
Y X
X