[English] 日本語
 Yorodumi
Yorodumi- EMDB-35301: Structure of mammalian spectrin-actin junctional complex of membr... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
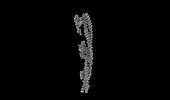
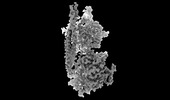
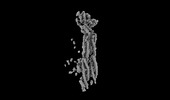
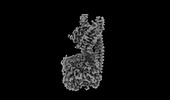
| Title | Structure of mammalian spectrin-actin junctional complex of membrane skeleton, State I, Global map | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationNCAM signaling for neurite out-growth / RAF/MAP kinase cascade / negative regulation of protein targeting to membrane / pointed-end actin filament capping / Gap junction degradation / Formation of annular gap junctions / Regulation of actin dynamics for phagocytic cup formation / EPHB-mediated forward signaling / VEGFA-VEGFR2 Pathway / Cell-extracellular matrix interactions ...NCAM signaling for neurite out-growth / RAF/MAP kinase cascade / negative regulation of protein targeting to membrane / pointed-end actin filament capping / Gap junction degradation / Formation of annular gap junctions / Regulation of actin dynamics for phagocytic cup formation / EPHB-mediated forward signaling / VEGFA-VEGFR2 Pathway / Cell-extracellular matrix interactions / RHO GTPases Activate WASPs and WAVEs / MAP2K and MAPK activation /  cuticular plate / cuticular plate /  spectrin / spectrin /  Clathrin-mediated endocytosis / lymphocyte homeostasis / lens fiber cell development / Clathrin-mediated endocytosis / lymphocyte homeostasis / lens fiber cell development /  myofibril assembly / spectrin-associated cytoskeleton / negative regulation of peptidyl-tyrosine phosphorylation / negative regulation of substrate adhesion-dependent cell spreading / porphyrin-containing compound biosynthetic process / cellular response to cytochalasin B / plasma membrane organization / platelet dense tubular network membrane / regulation of transepithelial transport / structural constituent of postsynaptic actin cytoskeleton / morphogenesis of a polarized epithelium / negative regulation of focal adhesion assembly / postsynaptic actin cytoskeleton / protein localization to adherens junction / cell projection membrane / dense body / Tat protein binding / myofibril assembly / spectrin-associated cytoskeleton / negative regulation of peptidyl-tyrosine phosphorylation / negative regulation of substrate adhesion-dependent cell spreading / porphyrin-containing compound biosynthetic process / cellular response to cytochalasin B / plasma membrane organization / platelet dense tubular network membrane / regulation of transepithelial transport / structural constituent of postsynaptic actin cytoskeleton / morphogenesis of a polarized epithelium / negative regulation of focal adhesion assembly / postsynaptic actin cytoskeleton / protein localization to adherens junction / cell projection membrane / dense body / Tat protein binding /  regulation of filopodium assembly / regulation of filopodium assembly /  COP9 signalosome / apical protein localization / actin filament capping / COP9 signalosome / apical protein localization / actin filament capping /  adherens junction assembly / RHO GTPases activate IQGAPs / RHO GTPases Activate Formins / adherens junction assembly / RHO GTPases activate IQGAPs / RHO GTPases Activate Formins /  regulation of lamellipodium assembly / positive regulation of fibroblast migration / regulation of lamellipodium assembly / positive regulation of fibroblast migration /  tight junction / regulation of norepinephrine uptake / COPI-mediated anterograde transport / tight junction / regulation of norepinephrine uptake / COPI-mediated anterograde transport /  NuA4 histone acetyltransferase complex / regulation of synaptic vesicle endocytosis / apical junction complex / erythrocyte development / establishment or maintenance of cell polarity / NuA4 histone acetyltransferase complex / regulation of synaptic vesicle endocytosis / apical junction complex / erythrocyte development / establishment or maintenance of cell polarity /  spectrin binding / spectrin binding /  tropomyosin binding / positive regulation of double-strand break repair via homologous recombination / cortical cytoskeleton / negative regulation of peptidyl-threonine phosphorylation / tropomyosin binding / positive regulation of double-strand break repair via homologous recombination / cortical cytoskeleton / negative regulation of peptidyl-threonine phosphorylation /  nitric-oxide synthase binding / striated muscle thin filament / nitric-oxide synthase binding / striated muscle thin filament /  brush border / brush border /  kinesin binding / kinesin binding /  calyx of Held / calyx of Held /  hemopoiesis / hemopoiesis /  : / positive regulation of blood coagulation / regulation of protein localization to plasma membrane / negative regulation of peptidyl-serine phosphorylation / cellular response to cAMP / positive regulation of T cell proliferation / cellular response to calcium ion / : / positive regulation of blood coagulation / regulation of protein localization to plasma membrane / negative regulation of peptidyl-serine phosphorylation / cellular response to cAMP / positive regulation of T cell proliferation / cellular response to calcium ion /  axonogenesis / adult locomotory behavior / axonogenesis / adult locomotory behavior /  muscle contraction / muscle contraction /  cell motility / cell motility /  actin filament / regulation of actin cytoskeleton organization / actin filament / regulation of actin cytoskeleton organization /  adherens junction / adherens junction /  Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / Schaffer collateral - CA1 synapse / structural constituent of cytoskeleton / cytoplasmic ribonucleoprotein granule / : / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / Schaffer collateral - CA1 synapse / structural constituent of cytoskeleton / cytoplasmic ribonucleoprotein granule / : /  nucleosome / nucleosome /  actin cytoskeleton / actin cytoskeleton /  lamellipodium / lamellipodium /  actin binding / positive regulation of protein binding / regulation of cell shape / cytoplasmic vesicle / actin cytoskeleton organization / protein-containing complex assembly / actin binding / positive regulation of protein binding / regulation of cell shape / cytoplasmic vesicle / actin cytoskeleton organization / protein-containing complex assembly /  postsynaptic density / postsynaptic density /  cytoskeleton / cytoskeleton /  regulation of cell cycle / regulation of cell cycle /  hydrolase activity / hydrolase activity /  ribonucleoprotein complex / ribonucleoprotein complex /  axon / axon /  signaling receptor binding / signaling receptor binding /  focal adhesion / focal adhesion /  synapse / glutamatergic synapse synapse / glutamatergic synapseSimilarity search - Function | |||||||||
| Biological species |   Sus scrofa (pig) / Sus scrofa (pig) /   pig (pig) pig (pig) | |||||||||
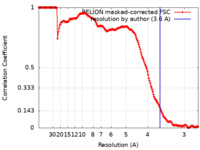
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.6 Å cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Li N / Chen S / Gao N | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: Structural basis of membrane skeleton organization in red blood cells. Authors: Ningning Li / Siyi Chen / Kui Xu / Meng-Ting He / Meng-Qiu Dong / Qiangfeng Cliff Zhang / Ning Gao /  Abstract: The spectrin-based membrane skeleton is a ubiquitous membrane-associated two-dimensional cytoskeleton underneath the lipid membrane of metazoan cells. Mutations of skeleton proteins impair the ...The spectrin-based membrane skeleton is a ubiquitous membrane-associated two-dimensional cytoskeleton underneath the lipid membrane of metazoan cells. Mutations of skeleton proteins impair the mechanical strength and functions of the membrane, leading to several different types of human diseases. Here, we report the cryo-EM structures of the native spectrin-actin junctional complex (from porcine erythrocytes), which is a specialized short F-actin acting as the central organizational unit of the membrane skeleton. While an α-/β-adducin hetero-tetramer binds to the barbed end of F-actin as a flexible cap, tropomodulin and SH3BGRL2 together create an absolute cap at the pointed end. The junctional complex is strengthened by ring-like structures of dematin in the middle actin layers and by patterned periodic interactions with tropomyosin over its entire length. This work serves as a structural framework for understanding the assembly and dynamics of membrane skeleton and offers insights into mechanisms of various ubiquitous F-actin-binding factors in other F-actin systems. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35301.map.gz emd_35301.map.gz | 398.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35301-v30.xml emd-35301-v30.xml emd-35301.xml emd-35301.xml | 27.4 KB 27.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35301_fsc.xml emd_35301_fsc.xml | 16.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_35301.png emd_35301.png | 36.7 KB | ||
| Others |  emd_35301_half_map_1.map.gz emd_35301_half_map_1.map.gz emd_35301_half_map_2.map.gz emd_35301_half_map_2.map.gz | 392 MB 392 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35301 http://ftp.pdbj.org/pub/emdb/structures/EMD-35301 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35301 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35301 | HTTPS FTP |
-Related structure data
| Related structure data |  8iahMC  8iaiC  8ib2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35301.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35301.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.37 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35301_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_35301_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Spectrin-actin junctional complex
+Supramolecule #1: Spectrin-actin junctional complex
+Macromolecule #1: Adducin 1
+Macromolecule #2: Beta-adducin
+Macromolecule #3: Dematin actin binding protein
+Macromolecule #4: Spectrin alpha, erythrocytic 1
+Macromolecule #5: Actin, cytoplasmic 1
+Macromolecule #6: Spectrin beta chain
+Macromolecule #7: Tropomyosin-1.9
+Macromolecule #8: Tropomyosin 3
+Macromolecule #9: Tropomodulin-1
+Macromolecule #10: SH3 domain-binding glutamic acid-rich-like protein
+Macromolecule #11: ADENOSINE-5'-DIPHOSPHATE
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 2.0 µm Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 2.0 µm |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 34.4 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z
Z Y
Y X
X