[English] 日本語
 Yorodumi
Yorodumi- EMDB-35085: Cryo-EM structure of Arabidopsis thaliana SOS1 in an occluded sta... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Arabidopsis thaliana SOS1 in an occluded state, with expanded TMD | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Sodium/proton antiporter /  MEMBRANE PROTEIN MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsodium:proton antiporter activity /  chloroplast envelope / regulation of reactive oxygen species metabolic process / sodium ion transport / potassium ion transmembrane transport / response to salt stress / response to reactive oxygen species / response to hydrogen peroxide / response to oxidative stress / chloroplast envelope / regulation of reactive oxygen species metabolic process / sodium ion transport / potassium ion transmembrane transport / response to salt stress / response to reactive oxygen species / response to hydrogen peroxide / response to oxidative stress /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Arabidopsis thaliana (thale cress) Arabidopsis thaliana (thale cress) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.4 Å cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Wang Y / Zhao Y / Gao Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Architecture and autoinhibitory mechanism of the plasma membrane Na/H antiporter SOS1 in Arabidopsis. Authors: Yuhang Wang / Chengcai Pan / Qihao Chen / Qing Xie / Yiwei Gao / Lingli He / Yue Li / Yanli Dong / Xingyu Jiang / Yan Zhao /  Abstract: Salt-overly-sensitive 1 (SOS1) is a unique electroneutral Na/H antiporter at the plasma membrane of higher plants and plays a central role in resisting salt stress. SOS1 is kept in a resting state ...Salt-overly-sensitive 1 (SOS1) is a unique electroneutral Na/H antiporter at the plasma membrane of higher plants and plays a central role in resisting salt stress. SOS1 is kept in a resting state with basal activity and activated upon phosphorylation. Here, we report the structures of SOS1. SOS1 forms a homodimer, with each monomer composed of transmembrane and intracellular domains. We find that SOS1 is locked in an occluded state by shifting of the lateral-gate TM5b toward the dimerization domain, thus shielding the Na/H binding site. We speculate that the dimerization of the intracellular domain is crucial to stabilize the transporter in this specific conformation. Moreover, two discrete fragments and a residue W1013 are important to prevent the transition of SOS1 to an alternative conformational state, as validated by functional complementation assays. Our study enriches understanding of the alternate access model of eukaryotic Na/H exchangers. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35085.map.gz emd_35085.map.gz | 116.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35085-v30.xml emd-35085-v30.xml emd-35085.xml emd-35085.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35085.png emd_35085.png | 138.5 KB | ||
| Filedesc metadata |  emd-35085.cif.gz emd-35085.cif.gz | 5.9 KB | ||
| Others |  emd_35085_half_map_1.map.gz emd_35085_half_map_1.map.gz emd_35085_half_map_2.map.gz emd_35085_half_map_2.map.gz | 115.6 MB 115.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35085 http://ftp.pdbj.org/pub/emdb/structures/EMD-35085 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35085 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35085 | HTTPS FTP |
-Related structure data
| Related structure data |  8hyaMC  7y3eC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35085.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35085.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||
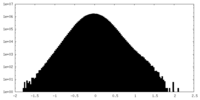
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35085_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
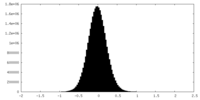
| Density Histograms |
-Half map: #1
| File | emd_35085_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
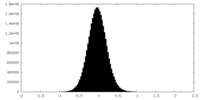
| Density Histograms |
- Sample components
Sample components
-Entire : Arabidopsis sodium/hydrogen exchanger 7 (SOS1), expand
| Entire | Name: Arabidopsis sodium/hydrogen exchanger 7 (SOS1), expand |
|---|---|
| Components |
|
-Supramolecule #1: Arabidopsis sodium/hydrogen exchanger 7 (SOS1), expand
| Supramolecule | Name: Arabidopsis sodium/hydrogen exchanger 7 (SOS1), expand type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Arabidopsis thaliana (thale cress) Arabidopsis thaliana (thale cress) |
-Macromolecule #1: Sodium/hydrogen exchanger 7
| Macromolecule | Name: Sodium/hydrogen exchanger 7 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Arabidopsis thaliana (thale cress) Arabidopsis thaliana (thale cress) |
| Molecular weight | Theoretical: 127.327891 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MTTVIDATMA YRFLEEATDS SSSSSSSKLE SSPVDAVLFV GMSLVLGIAS RHLLRGTRVP YTVALLVIGI ALGSLEYGAK HNLGKIGHG IRIWNEIDPE LLLAVFLPAL LFESSFSMEV HQIKRCLGQM VLLAVPGVLI STACLGSLVK VTFPYEWDWK T SLLLGGLL ...String: MTTVIDATMA YRFLEEATDS SSSSSSSKLE SSPVDAVLFV GMSLVLGIAS RHLLRGTRVP YTVALLVIGI ALGSLEYGAK HNLGKIGHG IRIWNEIDPE LLLAVFLPAL LFESSFSMEV HQIKRCLGQM VLLAVPGVLI STACLGSLVK VTFPYEWDWK T SLLLGGLL SATDPVAVVA LLKELGASKK LSTIIEGESL MNDGTAIVVF QLFLKMAMGQ NSDWSSIIKF LLKVALGAVG IG LAFGIAS VIWLKFIFND TVIEITLTIA VSYFAYYTAQ EWAGASGVLT VMTLGMFYAA FARTAFKGDS QKSLHHFWEM VAY IANTLI FILSGVVIAE GILDSDKIAY QGNSWRFLFL LYVYIQLSRV VVVGVLYPLL CRFGYGLDWK ESIILVWSGL RGAV ALALS LSVKQSSGNS HISKETGTLF LFFTGGIVFL TLIVNGSTTQ FVLRLLRMDI LPAPKKRILE YTKYEMLNKA LRAFQ DLGD DEELGPADWP TVESYISSLK GSEGELVHHP HNGSKIGSLD PKSLKDIRMR FLNGVQATYW EMLDEGRISE VTANIL MQS VDEALDQVST TLCDWRGLKP HVNFPNYYNF LHSKVVPRKL VTYFAVERLE SACYISAAFL RAHTIARQQL YDFLGES NI GSIVINESEK EGEEAKKFLE KVRSSFPQVL RVVKTKQVTY SVLNHLLGYI ENLEKVGLLE EKEIAHLHDA VQTGLKKL L RNPPIVKLPK LSDMITSHPL SVALPPAFCE PLKHSKKEPM KLRGVTLYKE GSKPTGVWLI FDGIVKWKSK ILSNNHSLH PTFSHGSTLG LYEVLTGKPY LCDLITDSMV LCFFIDSEKI LSLQSDSTID DFLWQESALV LLKLLRPQIF ESVAMQELRA LVSTESSKL TTYVTGESIE IDCNSIGLLL EGFVKPVGIK EELISSPAAL SPSNGNQSFH NSSEASGIMR VSFSQQATQY I VETRARAI IFNIGAFGAD RTLHRRPSSL TPPRSSSSDQ LQRSFRKEHR GLMSWPENIY AKQQQEINKT TLSLSERAMQ LS IFGSMVN VYRRSVSFGG IYNNKLQDNL LYKKLPLNPA QGLVSAKSES SIVTKKQLET RKHACQLPLK GESSTRQNTM VES SDEEDE DEGIVVRIDS PSKIVFRNDL UniProtKB: Sodium/hydrogen exchanger 7 |
-Macromolecule #2: HEXADECANE
| Macromolecule | Name: HEXADECANE / type: ligand / ID: 2 / Number of copies: 6 / Formula: R16 |
|---|---|
| Molecular weight | Theoretical: 226.441 Da |
| Chemical component information |  ChemComp-R16: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm Bright-field microscopy / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 31721 |
 Movie
Movie Controller
Controller







 Z
Z Y
Y X
X