+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Clr4-H3K9 Nucleosome complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | H3K9 nucleosome / methyltransferases / Clr4 /  PROTEIN BINDING PROTEIN BINDING | |||||||||
| Biological species |   Escherichia coli (E. coli) Escherichia coli (E. coli) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.5 Å cryo EM / Resolution: 4.5 Å | |||||||||
 Authors Authors | Akoury E | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2024 Journal: Sci Rep / Year: 2024Title: Structural insights into the binding mechanism of Clr4 methyltransferase to H3K9 methylated nucleosome. Authors: Christopher Saab / Joseph Stephan / Elias Akoury /  Abstract: The establishment and maintenance of heterochromatin, a specific chromatin structure essential for genomic stability and regulation, rely on intricate interactions between chromatin-modifying enzymes ...The establishment and maintenance of heterochromatin, a specific chromatin structure essential for genomic stability and regulation, rely on intricate interactions between chromatin-modifying enzymes and nucleosomal histone proteins. However, the precise trigger for these modifications remains unclear, thus highlighting the need for a deeper understanding of how methyltransferases facilitate histone methylation among others. Here, we investigate the molecular mechanisms underlying heterochromatin assembly by studying the interaction between the H3K9 methyltransferase Clr4 and H3K9-methylated nucleosomes. Using a combination of liquid-state nuclear magnetic resonance spectroscopy and cryo-electron microscopy, we elucidate the structural basis of Clr4 binding to H3K9-methylated nucleosomes. Our results reveal that Clr4 engages with nucleosomes through its chromodomain and disordered regions to promote de novo methylation. This study provides crucial insights into the molecular mechanisms governing heterochromatin formation by highlighting the significance of chromatin-modifying enzymes in genome regulation and disease pathology. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35060.map.gz emd_35060.map.gz | 1.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35060-v30.xml emd-35060-v30.xml emd-35060.xml emd-35060.xml | 11.2 KB 11.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35060.png emd_35060.png | 58.2 KB | ||
| Filedesc metadata |  emd-35060.cif.gz emd-35060.cif.gz | 3.7 KB | ||
| Others |  emd_35060_half_map_1.map.gz emd_35060_half_map_1.map.gz emd_35060_half_map_2.map.gz emd_35060_half_map_2.map.gz | 1.8 MB 1.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35060 http://ftp.pdbj.org/pub/emdb/structures/EMD-35060 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35060 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35060 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_35060.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35060.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_35060_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
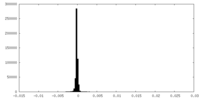
| Density Histograms |
-Half map: #2
| File | emd_35060_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
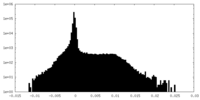
| Density Histograms |
- Sample components
Sample components
-Entire : Clr4-H3K9 Nucleosome complex
| Entire | Name: Clr4-H3K9 Nucleosome complex |
|---|---|
| Components |
|
-Supramolecule #1: Clr4-H3K9 Nucleosome complex
| Supramolecule | Name: Clr4-H3K9 Nucleosome complex / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Molecular weight | Theoretical: 230 KDa |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 |
|---|---|
| Grid | Material: COPPER |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.0 µm |
| Image recording | Film or detector model: TVIPS TEMCAM-F816 (8k x 8k) / Average electron dose: 15.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: COMMON LINE |
| Final reconstruction | Algorithm: ALGEBRAIC (ARTS) / Resolution.type: BY AUTHOR / Resolution: 4.5 Å / Resolution method: FSC 0.5 CUT-OFF / Number images used: 1200000 |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X