+ Open data
Open data
- Basic information
Basic information
| Entry | 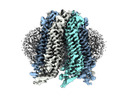 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human Pannexin-2 in pre-open state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Pannexin / Pannexin /  ATP / Inner channel lipids / ATP / Inner channel lipids /  Cell-cell communication / Cell-cell communication /  MEMBRANE PROTEIN MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationElectric Transmission Across Gap Junctions / wide pore channel activity / positive regulation of interleukin-1 production /  gap junction / monoatomic cation transport / monoatomic ion transmembrane transport / response to ischemia / cell-cell signaling / structural molecule activity / gap junction / monoatomic cation transport / monoatomic ion transmembrane transport / response to ischemia / cell-cell signaling / structural molecule activity /  plasma membrane / plasma membrane /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.4 Å cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Cong Y / Jin X / Kong F / Yan C | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Inner channel lipids regulated gating mechanism of human pannexins. Authors: Cong Y / Jin X / Kong F / Yan C | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34376.map.gz emd_34376.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34376-v30.xml emd-34376-v30.xml emd-34376.xml emd-34376.xml | 14.8 KB 14.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34376.png emd_34376.png | 133.9 KB | ||
| Masks |  emd_34376_msk_1.map emd_34376_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-34376.cif.gz emd-34376.cif.gz | 5.6 KB | ||
| Others |  emd_34376_half_map_1.map.gz emd_34376_half_map_1.map.gz emd_34376_half_map_2.map.gz emd_34376_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34376 http://ftp.pdbj.org/pub/emdb/structures/EMD-34376 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34376 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34376 | HTTPS FTP |
-Related structure data
| Related structure data |  8gyqMC  8gyoC  8gypC  8gytC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34376.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34376.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_34376_msk_1.map emd_34376_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_34376_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_34376_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Pannexin-2
| Entire | Name: Pannexin-2 |
|---|---|
| Components |
|
-Supramolecule #1: Pannexin-2
| Supramolecule | Name: Pannexin-2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 500 KDa |
-Macromolecule #1: Pannexin-2
| Macromolecule | Name: Pannexin-2 / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 74.54193 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MHHLLEQSAD MATALLAGEK LRELILPGAQ DDKAGALAAL LLQLKLELPF DRVVTIGTVL VPILLVTLVF TKNFAEEPIY CYTPHNFTR DQALYARGYC WTELRDALPG VDASLWPSLF EHKFLPYALL AFAAIMYVPA LGWEFLASTR LTSELNFLLQ E IDNCYHRA ...String: MHHLLEQSAD MATALLAGEK LRELILPGAQ DDKAGALAAL LLQLKLELPF DRVVTIGTVL VPILLVTLVF TKNFAEEPIY CYTPHNFTR DQALYARGYC WTELRDALPG VDASLWPSLF EHKFLPYALL AFAAIMYVPA LGWEFLASTR LTSELNFLLQ E IDNCYHRA AEGRAPKIEK QIQSKGPGIT EREKREIIEN AEKEKSPEQN LFEKYLERRG RSNFLAKLYL ARHVLILLLS AV PISYLCT YYATQKQNEF TCALGASPDG AAGAGPAVRV SCKLPSVQLQ RIIAGVDIVL LCVMNLIILV NLIHLFIFRK SNF IFDKLH KVGIKTRRQW RRSQFCDINI LAMFCNENRD HIKSLNRLDF ITNESDLMYD NVVRQLLAAL AQSNHDATPT VRDS GVQTV DPSANPAEPD GAAEPPVVKR PRKKMKWIPT SNPLPQPFKE PLAIMRVENS KAEKPKPARR KTATDTLIAP LLDRS AHHY KGGGGDPGPG PAPAPAPPPA PDKKHARHFS LDVHPYILGT KKAKAEAVPA ALPASRSQEG GFLSQAEDCG LGLAPA PIK DAPLPEKEIP YPTEPARAGL PSGGPFHVRS PPAAPAVAPL TPASLGKAEP LTILSRNATH PLLHINTLYE AREEEDG GP RLPQDVGDLI AIPAPQQILI ATFDEPRTVV STVEF UniProtKB:  Pannexin-2 Pannexin-2 |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 7 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.8 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 131444 |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)