[English] 日本語
 Yorodumi
Yorodumi- EMDB-33410: Deterget-solubilized E. coli RseP(L358C) mutant in complex with Fab -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Deterget-solubilized E. coli RseP(L358C) mutant in complex with Fab | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |   Escherichia coli (E. coli) / Escherichia coli (E. coli) /   Mus musculus (house mouse) Mus musculus (house mouse) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 14.0 Å negative staining / Resolution: 14.0 Å | |||||||||
 Authors Authors | Aruga R / Hirose M / Hirose T / Katagiri S / Iwasaki K / Kato T / Nogi T | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Mechanistic insights into intramembrane proteolysis by site-2 protease homolog RseP. Authors: Yuki Imaizumi / Kazunori Takanuki / Takuya Miyake / Mizuki Takemoto / Kunio Hirata / Mika Hirose / Rika Oi / Tatsuya Kobayashi / Kenichi Miyoshi / Rie Aruga / Tatsuhiko Yokoyama / Shizuka ...Authors: Yuki Imaizumi / Kazunori Takanuki / Takuya Miyake / Mizuki Takemoto / Kunio Hirata / Mika Hirose / Rika Oi / Tatsuya Kobayashi / Kenichi Miyoshi / Rie Aruga / Tatsuhiko Yokoyama / Shizuka Katagiri / Hiroaki Matsuura / Kenji Iwasaki / Takayuki Kato / Mika K Kaneko / Yukinari Kato / Michiko Tajiri / Satoko Akashi / Osamu Nureki / Yohei Hizukuri / Yoshinori Akiyama / Terukazu Nogi /  Abstract: Site-2 proteases are a conserved family of intramembrane proteases that cleave transmembrane substrates to regulate signal transduction and maintain proteostasis. Here, we elucidated crystal ...Site-2 proteases are a conserved family of intramembrane proteases that cleave transmembrane substrates to regulate signal transduction and maintain proteostasis. Here, we elucidated crystal structures of inhibitor-bound forms of bacterial site-2 proteases including RseP. Structure-based chemical modification and cross-linking experiments indicated that the RseP domains surrounding the active center undergo conformational changes to expose the substrate-binding site, suggesting that RseP has a gating mechanism to regulate substrate entry. Furthermore, mutational analysis suggests that a conserved electrostatic linkage between the transmembrane and peripheral membrane-associated domains mediates the conformational changes. In vivo cleavage assays also support that the substrate transmembrane helix is unwound by strand addition to the intramembrane β sheet of RseP and is clamped by a conserved asparagine residue at the active center for efficient cleavage. This mechanism underlying the substrate binding, i.e., unwinding and clamping, appears common across distinct families of intramembrane proteases that cleave transmembrane segments. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33410.map.gz emd_33410.map.gz | 12.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33410-v30.xml emd-33410-v30.xml emd-33410.xml emd-33410.xml | 27.2 KB 27.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33410.png emd_33410.png | 26.4 KB | ||
| Others |  emd_33410_additional_1.map.gz emd_33410_additional_1.map.gz emd_33410_additional_2.map.gz emd_33410_additional_2.map.gz emd_33410_additional_3.map.gz emd_33410_additional_3.map.gz emd_33410_additional_4.map.gz emd_33410_additional_4.map.gz emd_33410_half_map_1.map.gz emd_33410_half_map_1.map.gz emd_33410_half_map_2.map.gz emd_33410_half_map_2.map.gz | 12.4 MB 12.4 MB 12.4 MB 12.4 MB 65.7 MB 65.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33410 http://ftp.pdbj.org/pub/emdb/structures/EMD-33410 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33410 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33410 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33410.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33410.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.4 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
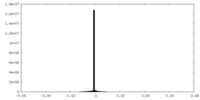
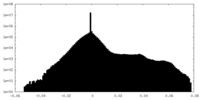
-Additional map: #1
| File | emd_33410_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
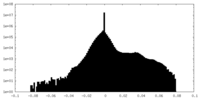
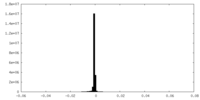
| Density Histograms |
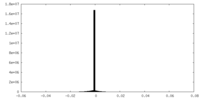
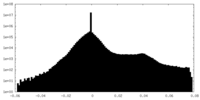
-Additional map: #2
| File | emd_33410_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
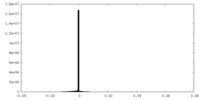
| Density Histograms |
-Additional map: #3
| File | emd_33410_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
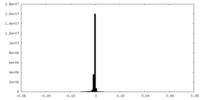
| Density Histograms |
-Additional map: #4
| File | emd_33410_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
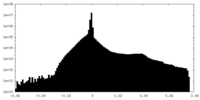
| Density Histograms |
-Half map: #1
| File | emd_33410_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_33410_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Binary complex of E. coli RseP(L358C) mutant with Fab
| Entire | Name: Binary complex of E. coli RseP(L358C) mutant with Fab |
|---|---|
| Components |
|
-Supramolecule #1: Binary complex of E. coli RseP(L358C) mutant with Fab
| Supramolecule | Name: Binary complex of E. coli RseP(L358C) mutant with Fab / type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: RseP
| Supramolecule | Name: RseP / type: complex / Chimera: Yes / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Supramolecule #3: Fab
| Supramolecule | Name: Fab / type: complex / Chimera: Yes / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
-Macromolecule #1: E. coli RseP(L358C) mutant
| Macromolecule | Name: E. coli RseP(L358C) mutant / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MLSFLWDLAS FIVALGVLIT VHGFGHFWV ARRCGVRVER F SIGFGKAL WRRTDKLGTE YV IALIPLG GYVKMLDERA EPV VPELRH HAFNNKSVGQ RAAI IAAGP VANFIFAIFA YWLVF IIGV PGVRPVVGEI AANSIA AEA QIAPGTELKA VDGIETP DW ...String: MLSFLWDLAS FIVALGVLIT VHGFGHFWV ARRCGVRVER F SIGFGKAL WRRTDKLGTE YV IALIPLG GYVKMLDERA EPV VPELRH HAFNNKSVGQ RAAI IAAGP VANFIFAIFA YWLVF IIGV PGVRPVVGEI AANSIA AEA QIAPGTELKA VDGIETP DW DAVRLQLVDK IGDESTTI T VAPFGSDQRR DVKLDLRHW AFEPDKEDPV SSLGIRPRGP QIEPVLENV QPNSAASKAG L QAGDRIVK VDGQPLTQWV TF VMLVRDN PGKSLALEIE RQG SPLSLT LIPESKPGNG KAIG FVGIE PKVIPLPDEY KVVRQ YGPF NAIVEATDKT WQLMKL TVS MLGKLITGDV KLNNCSG PI SIAKGAGMTA ELGVVYYL P FLALISVNLG IINLFPLPV LDGGHLLFLA IEKIKGGPVS ERVQDFCYR IGSILLVLLM G LALFNDFS RLGTENLYFQ |
-Macromolecule #2: Heavy chain of Fab
| Macromolecule | Name: Heavy chain of Fab / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Sequence | String: QVQLQQSRAE LARPGASVKM SCKASGYTFT TYTMQWVKQR P GQALEWIG YINPGSGYAK NNQKFKDKAT LTADKSSSTA YMQLSSLTSD DSAVYYCARS GS FFDYWGQ GTTLTVSSAK TTPPSVYPLA PGSAAQTNSM VTLGCLVKGY FPEPVTVTWN SGS LSSGVH ...String: QVQLQQSRAE LARPGASVKM SCKASGYTFT TYTMQWVKQR P GQALEWIG YINPGSGYAK NNQKFKDKAT LTADKSSSTA YMQLSSLTSD DSAVYYCARS GS FFDYWGQ GTTLTVSSAK TTPPSVYPLA PGSAAQTNSM VTLGCLVKGY FPEPVTVTWN SGS LSSGVH TFPAVLQSDL YTLSSSVTVP SSTWPSETVT CNVAHPASST KVDKKIVPRD C |
-Macromolecule #3: Light chain of Fab
| Macromolecule | Name: Light chain of Fab / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Sequence | String: DIVLTQSPAI LSVTPGDSVS LSCRASQSVS SNLHWYQQRS HESPRLLIT YAFQSISGIP SRFSGNGSGT DFTLNINSVE TEDFGMYFCQ QSNSWPYTFG G GTKLEIKR ADAAPTVSIF PPSSEQLTSG GASVVCFLNN FYPKDINVKW KIDGSERQNG VL NSWTDQD ...String: DIVLTQSPAI LSVTPGDSVS LSCRASQSVS SNLHWYQQRS HESPRLLIT YAFQSISGIP SRFSGNGSGT DFTLNINSVE TEDFGMYFCQ QSNSWPYTFG G GTKLEIKR ADAAPTVSIF PPSSEQLTSG GASVVCFLNN FYPKDINVKW KIDGSERQNG VL NSWTDQD SKDSTYSMSS TLTLTKDEYE RHNSYTCEAT HKTSTSPIVK SFNRNEC |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8.5 |
|---|---|
| Staining | Type: NEGATIVE / Material: Uranium accetate |
| Grid | Material: COPPER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 40.0 e/Å2 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 3279209 |
|---|---|
| CTF correction | Software - Name: CTFFIND (ver. 4.1) |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 3.1) |
| Final 3D classification | Number classes: 5 / Avg.num./class: 73152 / Software - Name: RELION (ver. 3.1) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 3.1) |
| Final reconstruction | Number classes used: 5 / Algorithm: FOURIER SPACE / Resolution.type: BY AUTHOR / Resolution: 14.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.1) / Number images used: 71271 |
 Movie
Movie Controller
Controller









 Z
Z Y
Y X
X