[English] 日本語
 Yorodumi
Yorodumi- EMDB-33385: Cryo-EM structure of the histone methyltransferase SET8 bound to ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 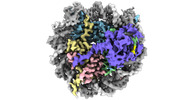 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the histone methyltransferase SET8 bound to H4K20Ecx-nucleosome | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationhistone H4K20 monomethyltransferase activity / lysine N-methyltransferase activity / [histone H4]-lysine20 N-methyltransferase / histone H4K20 methyltransferase activity / histone H4 methyltransferase activity / peptidyl-lysine monomethylation /  polytene chromosome / protein-lysine N-methyltransferase activity / mitotic chromosome condensation / regulation of DNA damage response, signal transduction by p53 class mediator ...histone H4K20 monomethyltransferase activity / lysine N-methyltransferase activity / [histone H4]-lysine20 N-methyltransferase / histone H4K20 methyltransferase activity / histone H4 methyltransferase activity / peptidyl-lysine monomethylation / polytene chromosome / protein-lysine N-methyltransferase activity / mitotic chromosome condensation / regulation of DNA damage response, signal transduction by p53 class mediator ...histone H4K20 monomethyltransferase activity / lysine N-methyltransferase activity / [histone H4]-lysine20 N-methyltransferase / histone H4K20 methyltransferase activity / histone H4 methyltransferase activity / peptidyl-lysine monomethylation /  polytene chromosome / protein-lysine N-methyltransferase activity / mitotic chromosome condensation / regulation of DNA damage response, signal transduction by p53 class mediator / polytene chromosome / protein-lysine N-methyltransferase activity / mitotic chromosome condensation / regulation of DNA damage response, signal transduction by p53 class mediator /  histone methyltransferase activity / negative regulation of double-strand break repair via homologous recombination / histone methyltransferase activity / negative regulation of double-strand break repair via homologous recombination /  Transferases; Transferring one-carbon groups; Methyltransferases / Condensation of Prophase Chromosomes / regulation of signal transduction by p53 class mediator / Regulation of TP53 Activity through Methylation / PKMTs methylate histone lysines / structural constituent of chromatin / transcription corepressor activity / Transferases; Transferring one-carbon groups; Methyltransferases / Condensation of Prophase Chromosomes / regulation of signal transduction by p53 class mediator / Regulation of TP53 Activity through Methylation / PKMTs methylate histone lysines / structural constituent of chromatin / transcription corepressor activity /  nucleosome / protein heterodimerization activity / nucleosome / protein heterodimerization activity /  cell division / negative regulation of DNA-templated transcription / cell division / negative regulation of DNA-templated transcription /  chromatin / regulation of transcription by RNA polymerase II / negative regulation of transcription by RNA polymerase II / chromatin / regulation of transcription by RNA polymerase II / negative regulation of transcription by RNA polymerase II /  DNA binding / DNA binding /  nucleoplasm / nucleoplasm /  nucleus / nucleus /  cytosol cytosolSimilarity search - Function | |||||||||
| Biological species |  Xenopus laevis (African clawed frog) / Xenopus laevis (African clawed frog) /   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.2 Å cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Shi LX / Zhou Z | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: FASEB J / Year: 2022 Journal: FASEB J / Year: 2022Title: Structural basis of nucleosomal H4K20 methylation by methyltransferase SET8. Authors: Liuxin Shi / Li Huang / Haizhen Long / Aoqun Song / Zheng Zhou /  Abstract: Histone H4 lysine 20 monomethylation (H4K20me1) plays a crucial role in multiple processes including DNA damage repair, DNA replication, and cell cycle control. Histone methyltransferase SET8 ...Histone H4 lysine 20 monomethylation (H4K20me1) plays a crucial role in multiple processes including DNA damage repair, DNA replication, and cell cycle control. Histone methyltransferase SET8 (previously named PR-Set7/KMT5A) mediates the chromatin deposition of H4K20me1, but how SET8 recognizes and modifies H4 in the context of the nucleosome is not fully understood. Here, we developed a simple chemical modification approach for H4K20 substitution by using the lysine analog S-ethyl-L-cysteine (Ecx). Substitution of H4K20 with H4Ecx20 improves the stability of the SET8-nucleosome complex, allowing us to determine the cryo-EM structure at 3.2 Å resolution. Structural analyses show that SET8 directly interacts with the H4 tail and the H2A-H2B acidic patch to ensure nucleosome binding. SET8 residues R339, K341, K351 make contact with nucleosomal DNA at the super helical location 2 (SHL2). Substitution of SET8 DNA-binding residues with alanines decreases the SET8-nucleosome interaction and impairs the methyltransferase activity. Disrupting the binding between SET8 R192 and H2A-H2B acidic patch decreases the cellular level of H4K20me1. Together, these results reveal a near-atomic resolution structure of SET8-bound nucleosome and provide insights into the SET8-mediated H4K20 recognition and modification. The lysine-to-Ecx substitution approach can be applied to the study of other methyltransferases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33385.map.gz emd_33385.map.gz | 60.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33385-v30.xml emd-33385-v30.xml emd-33385.xml emd-33385.xml | 22.9 KB 22.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33385.png emd_33385.png | 74.3 KB | ||
| Others |  emd_33385_half_map_1.map.gz emd_33385_half_map_1.map.gz emd_33385_half_map_2.map.gz emd_33385_half_map_2.map.gz | 49.7 MB 49.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33385 http://ftp.pdbj.org/pub/emdb/structures/EMD-33385 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33385 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33385 | HTTPS FTP |
-Related structure data
| Related structure data |  7xpxMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33385.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33385.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33385_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_33385_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Ternary complex of SET8 with H4K20Ecx nucleosome and SAM
+Supramolecule #1: Ternary complex of SET8 with H4K20Ecx nucleosome and SAM
+Supramolecule #2: Histone H3/H4/H2A/H2B
+Supramolecule #3: DNA
+Supramolecule #4: N-lysine methyltransferase KMT5A
+Macromolecule #1: Histone H3
+Macromolecule #2: Histone H4
+Macromolecule #3: Histone H2A
+Macromolecule #4: Histone H2B 1.1
+Macromolecule #7: N-lysine methyltransferase KMT5A
+Macromolecule #5: DNA (145-MER)
+Macromolecule #6: DNA (145-MER)
+Macromolecule #8: S-ADENOSYLMETHIONINE
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: DIFFRACTION / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.5 µm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.5 µm |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 2227867 |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.2 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 262534 |
 Movie
Movie Controller
Controller








 Z
Z Y
Y X
X



















