[English] 日本語
 Yorodumi
Yorodumi- EMDB-33225: Cryo-EM structure of SARS-CoV-2 prototype RBD-dimer bound to CB6 Fab -
+ Open data
Open data
- Basic information
Basic information
| Entry | 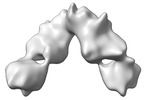 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
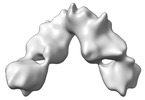
| Title | Cryo-EM structure of SARS-CoV-2 prototype RBD-dimer bound to CB6 Fab | |||||||||
 Map data Map data | Cryo-EM structure of SARS-CoV-2 prototype RBD-dimer bound to CB6 Fab | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Severe acute respiratory syndrome coronavirus 2 Severe acute respiratory syndrome coronavirus 2 | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 11.5 Å cryo EM / Resolution: 11.5 Å | |||||||||
 Authors Authors | George FG / Sheng L | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2022 Journal: Cell / Year: 2022Title: Protective prototype-Beta and Delta-Omicron chimeric RBD-dimer vaccines against SARS-CoV-2. Authors: Kun Xu / Ping Gao / Sheng Liu / Shuaiyao Lu / Wenwen Lei / Tianyi Zheng / Xueyuan Liu / Yufeng Xie / Zhennan Zhao / Shuxin Guo / Cong Tang / Yun Yang / Wenhai Yu / Junbin Wang / Yanan Zhou / ...Authors: Kun Xu / Ping Gao / Sheng Liu / Shuaiyao Lu / Wenwen Lei / Tianyi Zheng / Xueyuan Liu / Yufeng Xie / Zhennan Zhao / Shuxin Guo / Cong Tang / Yun Yang / Wenhai Yu / Junbin Wang / Yanan Zhou / Qing Huang / Chuanyu Liu / Yaling An / Rong Zhang / Yuxuan Han / Minrun Duan / Shaofeng Wang / Chenxi Yang / Changwei Wu / Xiaoya Liu / Guangbiao She / Yan Liu / Xin Zhao / Ke Xu / Jianxun Qi / Guizhen Wu / Xiaozhong Peng / Lianpan Dai / Peiyi Wang / George F Gao /  Abstract: Breakthrough infections by SARS-CoV-2 variants become the global challenge for pandemic control. Previously, we developed the protein subunit vaccine ZF2001 based on the dimeric receptor-binding ...Breakthrough infections by SARS-CoV-2 variants become the global challenge for pandemic control. Previously, we developed the protein subunit vaccine ZF2001 based on the dimeric receptor-binding domain (RBD) of prototype SARS-CoV-2. Here, we developed a chimeric RBD-dimer vaccine approach to adapt SARS-CoV-2 variants. A prototype-Beta chimeric RBD-dimer was first designed to adapt the resistant Beta variant. Compared with its homotypic forms, the chimeric vaccine elicited broader sera neutralization of variants and conferred better protection in mice. The protection of the chimeric vaccine was further verified in macaques. This approach was generalized to develop Delta-Omicron chimeric RBD-dimer to adapt the currently prevalent variants. Again, the chimeric vaccine elicited broader sera neutralization of SARS-CoV-2 variants and conferred better protection against challenge by either Delta or Omicron SARS-CoV-2 in mice. The chimeric approach is applicable for rapid updating of immunogens, and our data supported the use of variant-adapted multivalent vaccine against circulating and emerging variants. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33225.map.gz emd_33225.map.gz | 115.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33225-v30.xml emd-33225-v30.xml emd-33225.xml emd-33225.xml | 13.1 KB 13.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33225.png emd_33225.png | 35.8 KB | ||
| Others |  emd_33225_half_map_1.map.gz emd_33225_half_map_1.map.gz emd_33225_half_map_2.map.gz emd_33225_half_map_2.map.gz | 115.6 MB 115.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33225 http://ftp.pdbj.org/pub/emdb/structures/EMD-33225 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33225 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33225 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33225.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33225.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of SARS-CoV-2 prototype RBD-dimer bound to CB6 Fab | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.66 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Cryo-EM structure of SARS-CoV-2 prototype RBD-dimer bound to CB6 Fab
| File | emd_33225_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of SARS-CoV-2 prototype RBD-dimer bound to CB6 Fab | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Cryo-EM structure of SARS-CoV-2 prototype RBD-dimer bound to CB6 Fab
| File | emd_33225_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of SARS-CoV-2 prototype RBD-dimer bound to CB6 Fab | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of SARS-CoV-2 prototype RBD-dimer bound to CB6 Fab
| Entire | Name: Cryo-EM structure of SARS-CoV-2 prototype RBD-dimer bound to CB6 Fab |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of SARS-CoV-2 prototype RBD-dimer bound to CB6 Fab
| Supramolecule | Name: Cryo-EM structure of SARS-CoV-2 prototype RBD-dimer bound to CB6 Fab type: complex / ID: 1 / Chimera: Yes / Parent: 0 |
|---|---|
| Source (natural) | Organism:   Severe acute respiratory syndrome coronavirus 2 Severe acute respiratory syndrome coronavirus 2 |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.2 µm Bright-field microscopy / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.2 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 1.39 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Initial angle assignment | Type: OTHER |
|---|---|
| Final angle assignment | Type: OTHER |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 11.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 26842 |
 Movie
Movie Controller
Controller


 Z
Z Y
Y X
X