+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
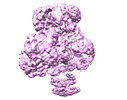
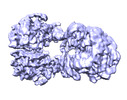
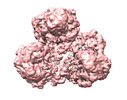
| Title | PC-(acetyl-CoA) (12.5uM) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.9 Å cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Chai P / Lan P / Wu J / Lei M | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2022 Journal: Mol Cell / Year: 2022Title: Mechanistic insight into allosteric activation of human pyruvate carboxylase by acetyl-CoA. Authors: Peiwei Chai / Pengfei Lan / Shaobai Li / Deqiang Yao / Chenchen Chang / Mi Cao / Yafeng Shen / Shengfang Ge / Jian Wu / Ming Lei / Xianqun Fan /  Abstract: Pyruvate carboxylase (PC) catalyzes the two-step carboxylation of pyruvate to produce oxaloacetate, playing a key role in the maintenance of metabolic homeostasis in cells. Given its involvement in ...Pyruvate carboxylase (PC) catalyzes the two-step carboxylation of pyruvate to produce oxaloacetate, playing a key role in the maintenance of metabolic homeostasis in cells. Given its involvement in multiple diseases, PC has been regarded as a potential therapeutic target for obesity, diabetes, and cancer. Albeit acetyl-CoA has been recognized as the allosteric regulator of PC for over 60 years, the underlying mechanism of how acetyl-CoA induces PC activation remains enigmatic. Herein, by using time-resolved cryo-electron microscopy, we have captured the snapshots of PC transitional states during its catalytic cycle. These structures and the biochemical studies reveal that acetyl-CoA stabilizes PC in a catalytically competent conformation, which triggers a cascade of events, including ATP hydrolysis and the long-distance communication between the two reactive centers. These findings provide an integrated picture for PC catalysis and unveil the unique allosteric mechanism of acetyl-CoA in an essential biochemical reaction in all kingdoms of life. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32774.map.gz emd_32774.map.gz | 5.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32774-v30.xml emd-32774-v30.xml emd-32774.xml emd-32774.xml | 9.7 KB 9.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32774.png emd_32774.png | 109.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32774 http://ftp.pdbj.org/pub/emdb/structures/EMD-32774 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32774 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32774 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32774.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32774.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : PC
| Entire | Name: PC |
|---|---|
| Components |
|
-Supramolecule #1: PC
| Supramolecule | Name: PC / type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Pyruvate carboxylase
| Macromolecule | Name: Pyruvate carboxylase / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MLKFRTVHGG LRLLGIRRTS TAPAASPNVR RLEYKPIKKV MVANRGEIAI RVFRACTELG IRTVAIYSEQ DTGQMHRQK ADEAYLIGRG LAPVQAYLHI PDIIKVAKEN NVDAVHPGYG FLSERADFAQ ACQDAGVRFI G PSPEVVRK MGDKVEARAI AIAAGVPVVP ...String: MLKFRTVHGG LRLLGIRRTS TAPAASPNVR RLEYKPIKKV MVANRGEIAI RVFRACTELG IRTVAIYSEQ DTGQMHRQK ADEAYLIGRG LAPVQAYLHI PDIIKVAKEN NVDAVHPGYG FLSERADFAQ ACQDAGVRFI G PSPEVVRK MGDKVEARAI AIAAGVPVVP GTDAPITSLH EAHEFSNTYG FPIIFKAAYG GGGRGMRVVH SY EELEENY TRAYSEALAA FGNGALFVEK FIEKPRHIEV QILGDQYGNI LHLYERDCSI QRRHQKVVEI APA AHLDPQ LRTRLTSDSV KLAKQVGYEN AGTVEFLVDR HGKHYFIEVN SRLQVEHTVT EEITDVDLVH AQIH VAEGR SLPDLGLRQE NIRINGCAIQ CRVTTEDPAR SFQPDTGRIE VFRSGEGMGI RLDNASAFQG AVISP HYDS LLVKVIAHGK DHPTAATKMS RALAEFRVRG VKTNIAFLQN VLNNQQFLAG TVDTQFIDEN PELFQL RPA QNRAQKLLHY LGHVMVNGPT TPIPVKASPS PTDPVVPAVP IGPPPAGFRD ILLREGPEGF ARAVRNH PG LLLMDTTFRD AHQSLLATRV RTHDLKKIAP YVAHNFSKLF SMENWGGATF DVAMRFLYEC PWRRLQEL R ELIPNIPFQM LLRGANAVGY TNYPDNVVFK FCEVAKENGM DVFRVFDSLN YLPNMLLGME AAGSAGGVV EAAISYTGDV ADPSRTKYSL QYYMGLAEEL VRAGTHILCI KDMAGLLKPT ACTMLVSSLR DRFPDLPLHI HTHDTSGAG VAAMLACAQA GADVVDVAAD SMSGMTSQPS MGALVACTRG TPLDTEVPME RVFDYSEYWE G ARGLYAAF DCTATMKSGN SDVYENEIPG GQYTNLHFQA HSMGLGSKFK EVKKAYVEAN QMLGDLIKVT PS SKIVGDL AQFMVQNGLS RAEAEAQAEE LSFPRSVVEF LQGYIGVPHG GFPEPFRSKV LKDLPRVEGR PGA SLPPLD LQALEKELVD RHGEEVTPED VLSAAMYPDV FAHFKDFTAT FGPLDSLNTR LFLQGPKIAE EFEV ELERG KTLHIKALAV SDLNRAGQRQ VFFELNGQLR SILVKDTQAM KEMHFHPKAL KDVKGQIGAP MPGKV IDIK VVAGAKVAKG QPLCVLSAMK METVVTSPME GTVRKVHVTK DMTLEGDDLI LEIE |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Initial angle assignment | Type: COMMON LINE |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 86474 |
 Movie
Movie Controller
Controller