+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
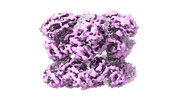
| Title | Cryo-EM structure of substrate engaged Drg1 hexamer | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |   Saccharomyces cerevisiae (brewer's yeast) / Saccharomyces cerevisiae (brewer's yeast) /   Escherichia coli (E. coli) Escherichia coli (E. coli) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.6 Å cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Ma CY / Wu DM / Chen Q / Gao N | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition. Authors: Chengying Ma / Damu Wu / Qian Chen / Ning Gao /  Abstract: The type II AAA + ATPase Drg1 is a ribosome assembly factor, functioning to release Rlp24 from the pre-60S particle just exported from nucleus, and its activity in can be inhibited by a drug ...The type II AAA + ATPase Drg1 is a ribosome assembly factor, functioning to release Rlp24 from the pre-60S particle just exported from nucleus, and its activity in can be inhibited by a drug molecule diazaborine. However, molecular mechanisms of Drg1-mediated Rlp24 removal and diazaborine-mediated inhibition are not fully understood. Here, we report Drg1 structures in different nucleotide-binding and benzo-diazaborine treated states. Drg1 hexamers transits between two extreme conformations (planar or helical arrangement of protomers). By forming covalent adducts with ATP molecules in both ATPase domain, benzo-diazaborine locks Drg1 hexamers in a symmetric and non-productive conformation to inhibits both inter-protomer and inter-ring communication of Drg1 hexamers. We also obtained a substrate-engaged mutant Drg1 structure, in which conserved pore-loops form a spiral staircase to interact with the polypeptide through a sequence-independent manner. Structure-based mutagenesis data highlight the functional importance of the pore-loop, the D1-D2 linker and the inter-subunit signaling motif of Drg1, which share similar regulatory mechanisms with p97. Our results suggest that Drg1 may function as an unfoldase that threads a substrate protein within the pre-60S particle. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32396.map.gz emd_32396.map.gz | 7.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32396-v30.xml emd-32396-v30.xml emd-32396.xml emd-32396.xml | 10.9 KB 10.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32396.png emd_32396.png | 65.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32396 http://ftp.pdbj.org/pub/emdb/structures/EMD-32396 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32396 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32396 | HTTPS FTP |
-Related structure data
| Related structure data |  7wbbMC  7wd3C  7ykkC  7yklC  7yktC  7ykzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32396.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32396.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.057 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Double mutant Drg1 in the presence of ATP with substrate engaged
| Entire | Name: Double mutant Drg1 in the presence of ATP with substrate engaged |
|---|---|
| Components |
|
-Supramolecule #1: Double mutant Drg1 in the presence of ATP with substrate engaged
| Supramolecule | Name: Double mutant Drg1 in the presence of ATP with substrate engaged type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) |
-Macromolecule #1: AFG2 isoform 1
| Macromolecule | Name: AFG2 isoform 1 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) |
| Molecular weight | Theoretical: 84.848742 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MAPKSSSSGS KKKSSASSNS ADAKASKFKL PAEFITRPHP SKDHGKETCT AYIHPNVLSS LEINPGSFCT VGKIGENGIL VIARAGDEE VHPVNVITLS TTIRSVGNLI LGDRLELKKA QVQPPYATKV TVGSLQGYNI LECMEEKVIQ KLLDDSGVIM P GMIFQNLK ...String: MAPKSSSSGS KKKSSASSNS ADAKASKFKL PAEFITRPHP SKDHGKETCT AYIHPNVLSS LEINPGSFCT VGKIGENGIL VIARAGDEE VHPVNVITLS TTIRSVGNLI LGDRLELKKA QVQPPYATKV TVGSLQGYNI LECMEEKVIQ KLLDDSGVIM P GMIFQNLK TKAGDESIDV VITDASDDSL PDVSQLDLNM DDMYGGLDNL FYLSPPFIFR KGSTHITFSK ETQANRKYNL PE PLSYAAV GGLDKEIESL KSAIEIPLHQ PTLFSSFGVS PPRGILLHGP PGTGKTMLLR VVANTSNAHV LTINGPSIVS KYL GETEAA LRDIFNEARK YQPSIIFIDQ IDSIAPNRAN DDSGEVESRV VATLLTLMDG MGAAGKVVVI AATNRPNSVD PALR RPGRF DQEVEIGIPD VDARFDILTK QFSRMSSDRH VLDSEAIKYI ASKTHGYVGA DLTALCRESV MKTIQRGLGT DANID KFSL KVTLKDVESA MVDIRPSAMR EIFLEMPKVY WSDIGGQEEL KTKMKEMIQL PLEASETFAR LGISAPKGVL LYGPPG CSK TLTAKALATE SGINFLAVKG PEIFNKYVGE SERAIREIFR KARSAAPSII FFDQIDALSP DRDGSSTSAA NHVLTSL LN EIDGVEELKG VVIVAATNRP DEIDAALLRP GRLDRHIYVG PPDVNARLEI LKKCTKKFNT EESGVDLHEL ADRTEGYS G AEVVLLCQEA GLAAIMEDLD VAKVELRHFE KAFKGIARGI TPEMLSYYEE FALRSGSSS |
-Macromolecule #2: substrate
| Macromolecule | Name: substrate / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Molecular weight | Theoretical: 1.975426 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) |
-Macromolecule #3: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 11 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 8.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
|---|---|
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.6 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 152973 |
 Movie
Movie Controller
Controller