+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Ams1 bound to the FW domain of Nbr1 | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
| Function / homology |  Function and homology information Function and homology information alpha-mannosidase / alpha-mannosidase /  alpha-mannosidase activity / mannose metabolic process / detection of maltose stimulus / alpha-mannosidase activity / mannose metabolic process / detection of maltose stimulus /  maltose binding / maltose transport complex / maltose transport / maltodextrin transmembrane transport / carbohydrate transmembrane transporter activity / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing ... maltose binding / maltose transport complex / maltose transport / maltodextrin transmembrane transport / carbohydrate transmembrane transporter activity / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing ... alpha-mannosidase / alpha-mannosidase /  alpha-mannosidase activity / mannose metabolic process / detection of maltose stimulus / alpha-mannosidase activity / mannose metabolic process / detection of maltose stimulus /  maltose binding / maltose transport complex / maltose transport / maltodextrin transmembrane transport / carbohydrate transmembrane transporter activity / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / carbohydrate transport / ATP-binding cassette (ABC) transporter complex / cell chemotaxis / outer membrane-bounded periplasmic space / maltose binding / maltose transport complex / maltose transport / maltodextrin transmembrane transport / carbohydrate transmembrane transporter activity / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / carbohydrate transport / ATP-binding cassette (ABC) transporter complex / cell chemotaxis / outer membrane-bounded periplasmic space /  carbohydrate binding / carbohydrate binding /  periplasmic space / DNA damage response / zinc ion binding / periplasmic space / DNA damage response / zinc ion binding /  membrane / membrane /  metal ion binding metal ion bindingSimilarity search - Function | ||||||||||||
| Biological species |  Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) / Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) /   Escherichia coli K-12 (bacteria) Escherichia coli K-12 (bacteria) | ||||||||||||
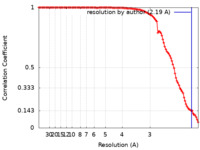
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.19 Å cryo EM / Resolution: 2.19 Å | ||||||||||||
 Authors Authors | Zhang J / Ye K | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural mechanism of protein recognition by the FW domain of autophagy receptor Nbr1 Authors: Zhang J / Wang YY / Pan ZQ / Li Y / Sui J / Du LL / Ye K | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32091.map.gz emd_32091.map.gz | 4.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32091-v30.xml emd-32091-v30.xml emd-32091.xml emd-32091.xml | 20.7 KB 20.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32091_fsc.xml emd_32091_fsc.xml | 11.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_32091.png emd_32091.png | 179.4 KB | ||
| Masks |  emd_32091_msk_1.map emd_32091_msk_1.map | 125 MB |  Mask map Mask map | |
| Others |  emd_32091_half_map_1.map.gz emd_32091_half_map_1.map.gz emd_32091_half_map_2.map.gz emd_32091_half_map_2.map.gz | 115.1 MB 115.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32091 http://ftp.pdbj.org/pub/emdb/structures/EMD-32091 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32091 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32091 | HTTPS FTP |
-Related structure data
| Related structure data |  7vqoMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32091.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32091.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_32091_msk_1.map emd_32091_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_32091_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_32091_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ams1, Nbr1 and malE fusion protein
| Entire | Name: Ams1, Nbr1 and malE fusion protein |
|---|---|
| Components |
|
-Supramolecule #1: Ams1, Nbr1 and malE fusion protein
| Supramolecule | Name: Ams1, Nbr1 and malE fusion protein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: The fusion protein comprises of the full-length Ams1, a linker sequence (GGGGSGGGFKKASSSDNKEQGGGGSGGGSG), residues 635-775 of Nbr1, and maltose binding protein (MBP). |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) Chaetomium thermophilum var. thermophilum DSM 1495 (fungus)Strain: DSM 1495 |
| Recombinant expression | Organism:   Schizosaccharomyces pombe 972h- (yeast) Schizosaccharomyces pombe 972h- (yeast) |
| Molecular weight | Theoretical: 520 KDa |
-Macromolecule #1: Ams1, Nbr1 and malE fusion protein
| Macromolecule | Name: Ams1, Nbr1 and malE fusion protein / type: protein_or_peptide / ID: 1 Details: The fusion protein comprises of the full-length Ams1, a linker sequence (GGGGSGGGFKKASSSDNKEQGGGGSGGGSG), residues 635-775 of Nbr1, and maltose binding protein (MBP).,The fusion protein ...Details: The fusion protein comprises of the full-length Ams1, a linker sequence (GGGGSGGGFKKASSSDNKEQGGGGSGGGSG), residues 635-775 of Nbr1, and maltose binding protein (MBP).,The fusion protein comprises of the full-length Ams1, a linker sequence (GGGGSGGGFKKASSSDNKEQGGGGSGGGSG), residues 635-775 of Nbr1, and maltose binding protein (MBP). Number of copies: 4 / Enantiomer: LEVO / EC number:  alpha-mannosidase alpha-mannosidase |
|---|---|
| Source (natural) | Organism:   Escherichia coli K-12 (bacteria) / Strain: K-12 Escherichia coli K-12 (bacteria) / Strain: K-12 |
| Molecular weight | Theoretical: 181.67625 KDa |
| Recombinant expression | Organism:   Schizosaccharomyces pombe 972h- (yeast) Schizosaccharomyces pombe 972h- (yeast) |
| Sequence | String: MGGETFDVKG PRPNDYPLRA PKPVGQLISH IYKDRIAQFY NGGQYEHQNL RAMMKEDSVS GEPHVQLWVW HAPGQTRPSF EEAVSNQFV KTNVGEWFGP SWTTHWFRVV LTVPEHLQNK RLLEFHWDSN SEGLVWSEDG KPLQGLTGGG ERVEWILPDS F RDGKEHTI ...String: MGGETFDVKG PRPNDYPLRA PKPVGQLISH IYKDRIAQFY NGGQYEHQNL RAMMKEDSVS GEPHVQLWVW HAPGQTRPSF EEAVSNQFV KTNVGEWFGP SWTTHWFRVV LTVPEHLQNK RLLEFHWDSN SEGLVWSEDG KPLQGLTGGG ERVEWILPDS F RDGKEHTI YIEMACNRMF GNAPGGDSIQ PPDPNKYFRL DKAEIVAIDP DARQLWIDIW ILQDAAREFP GDSWESHKAL QV CNEIIEA FELGNRESLK KCRKIAEQYL GPNVDSPNVY NSGKEPLVYA IGHCHIDSCW LWPFAETKRK VVRSWSSQCD LMD RYPELN FVCSQAQQYK WLKQLYPYAF ERVKKKVAEG RFHPIGGSWV EHDTNMPSGE SLVRQFLYGQ RFYESNFGKR CKTF WLPDT FGYSAQLPQL CRLAGMTRFL TQKLSWNNIN RFPHTTFNWV ALDGSQVICH MPPSETYTAE AHFGDVKRSM SQHKS LDQD NTSLLVFGKG DGGGGPTWVQ IEKLRRCRGI SDTVGLLPRV HMGSSVDDFF DRLERKADTF VTWYGELYFE LHRGTY TTQ AKNKKNNRRA EAKLRDLELL ATIASVQDKS YKYPKEEFDA MWENVLLCQF HDCLPGSSIE MAYRESDQMY ADVFSTA EK IMKGVSQVLG LEPALNHMST TNTVALNTLP WPRRELVKIS EKEAAVAHGT GPFLKLQKLE TTKPLVTLRQ VTKGAFVL E NSQLRVHVEK GVITSLYDKQ ANREVIPKGQ KANQYVIFDD KPLYWQAWDV EVYHLDTRKE LPSGETEVHE NTPHRVSVV TRTKVSDKSH IQTIIALNGA VEGEQSWVEV QSKVDWHETM KFLKVEFPVD VRNTEASYET AFGIVRRPTH YNTSWDMAKF EVCAHRWAD LSEYGYGVSI LNDSKYGFAT AGQTMRLSLL RSPKAPDAHA DMGTHHIRWA ILPHQGSLSH VTIRKAFEFN N PTKLYSSP DAAALVAAPP PVWLTPDSSP AIVLDTVKRG EDDEDVSRGE LPARKGQSVI LRMYDSLGGL ARGTVVTTWP LK KVCKVNL LEDDLEVVPW ENGRFTVELR PFEVASYRLV LAGGGGSGGG FKKASSSDNK EQGGGGSGGG SGEPVVEKEP SAE ELEATF VRDTVQDGTV LAPNHLFEQT WVLRNTGKVA WPAGCSVKFV GGDYMGRVDS AHPAASKEVE ESCESTVCDR AVQP GEEAP FTVLLRTPYR ACRVISHWRL TTPKGTKFGH RLWCDVVVEK PKSRSKIEEG KLVIWINGDK GYNGLAEVGK KFEKD TGIK VTVEHPDKLE EKFPQVAATG DGPDIIFWAH DRFGGYAQSG LLAEITPDKA FQDKLYPFTW DAVRYNGKLI AYPIAV EAL SLIYNKDLLP NPPKTWEEIP ALDKELKAKG KSALMFNLQE PYFTWPLIAA DGGYAFKYEN GKYDIKDVGV DNAGAKA GL TFLVDLIKNK HMNADTDYSI AEAAFNKGET AMTINGPWAW SNIDTSKVNY GVTVLPTFKG QPSKPFVGVL SAGINAAS P NKELAKEFLE NYLLTDEGLE AVNKDKPLGA VALKSYEEEL AKDPRIAATM ENAQKGEIMP NIPQMSAFWY AVRTAVINA ASGRQTVDEA LKDAQT |
-Macromolecule #2: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 2 / Number of copies: 4 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 688 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Homemade / Material: NICKEL/TITANIUM / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 3 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.5 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.5 µm |
| Specialist optics | Energy filter - Name: GIF Tridiem 4K / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number real images: 2925 / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z
Z Y
Y X
X