[English] 日本語
 Yorodumi
Yorodumi- EMDB-31964: Cryo-EM structure of Arabidopsis DCL3 in complex with a 30-bp RNA -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31964 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Arabidopsis DCL3 in complex with a 30-bp RNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationregulatory ncRNA-mediated post-transcriptional gene silencing /  ribonuclease III activity / ribonuclease III activity /  RNA processing / defense response to virus / RNA processing / defense response to virus /  nucleic acid binding / nucleic acid binding /  ATP binding / ATP binding /  nucleus nucleusSimilarity search - Function | |||||||||
| Biological species |   Arabidopsis thaliana (thale cress) Arabidopsis thaliana (thale cress) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.73 Å cryo EM / Resolution: 3.73 Å | |||||||||
 Authors Authors | Wang Q / Du J | |||||||||
 Citation Citation |  Journal: Science / Year: 2021 Journal: Science / Year: 2021Title: Mechanism of siRNA production by a plant Dicer-RNA complex in dicing-competent conformation. Authors: Qian Wang / Yan Xue / Laixing Zhang / Zhenhui Zhong / Suhua Feng / Changshi Wang / Lifan Xiao / Zhenlin Yang / C Jake Harris / Zhe Wu / Jixian Zhai / Maojun Yang / Sisi Li / Steven E Jacobsen / Jiamu Du /    Abstract: In eukaryotes, small RNAs (sRNAs) play critical roles in multiple biological processes. Dicer endonucleases are a central part of sRNA biogenesis. In plants, DICER-LIKE PROTEIN 3 (DCL3) produces 24- ...In eukaryotes, small RNAs (sRNAs) play critical roles in multiple biological processes. Dicer endonucleases are a central part of sRNA biogenesis. In plants, DICER-LIKE PROTEIN 3 (DCL3) produces 24-nucleotide (nt) small interfering RNAs (siRNAs) that determine the specificity of the RNA-directed DNA methylation pathway. Here, we determined the structure of a DCL3–pre-siRNA complex in an active dicing-competent state. The 5′-phosphorylated A1 of the guide strand and the 1-nt 3′ overhang of the complementary strand are specifically recognized by a positively charged pocket and an aromatic cap, respectively. The 24-nt siRNA length dependence relies on the separation between the 5′-phosphorylated end of the guide RNA and dual cleavage sites formed by the paired ribonuclease III domains. These structural studies, complemented by functional data, provide insight into the dicing principle for Dicers in general. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31964.map.gz emd_31964.map.gz | 5.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31964-v30.xml emd-31964-v30.xml emd-31964.xml emd-31964.xml | 16.3 KB 16.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31964.png emd_31964.png | 168.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31964 http://ftp.pdbj.org/pub/emdb/structures/EMD-31964 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31964 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31964 | HTTPS FTP |
-Related structure data
| Related structure data | 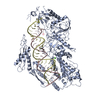 7vg3MC  7vg2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31964.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31964.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : DCL3 in complex with a 30-bp RNA
| Entire | Name: DCL3 in complex with a 30-bp RNA |
|---|---|
| Components |
|
-Supramolecule #1: DCL3 in complex with a 30-bp RNA
| Supramolecule | Name: DCL3 in complex with a 30-bp RNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Molecular weight | Experimental: 200 KDa |
-Supramolecule #2: DCL3
| Supramolecule | Name: DCL3 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Arabidopsis thaliana (thale cress) Arabidopsis thaliana (thale cress) |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
-Supramolecule #3: RNA
| Supramolecule | Name: RNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|
-Macromolecule #1: Dicer-like 3
| Macromolecule | Name: Dicer-like 3 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Arabidopsis thaliana (thale cress) Arabidopsis thaliana (thale cress) |
| Molecular weight | Theoretical: 182.176766 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MWSHPQFEKG GGARGGSGGG SWSHPQFEKG FDYKDDDDKG SGSENLYFQG SMHSSLEPEK MEEGGGSNSL KRKFSEIDGD QNLDSVSSP MMTDSNGSYE LKVYEVAKNR NIIAVLGTGI DKSEITKRLI KAMGSSDTDK RLIIFLAPTV NLVKQQCCEI R ALVNLKVE ...String: MWSHPQFEKG GGARGGSGGG SWSHPQFEKG FDYKDDDDKG SGSENLYFQG SMHSSLEPEK MEEGGGSNSL KRKFSEIDGD QNLDSVSSP MMTDSNGSYE LKVYEVAKNR NIIAVLGTGI DKSEITKRLI KAMGSSDTDK RLIIFLAPTV NLVKQQCCEI R ALVNLKVE EYFGAKGVDK WTSQRWDEEF SKHDVLVMTP QILLDVLRSA FLKLEMVCLL IIDECHHTTG NHPYAKLMKE FY HESTSKP KIFGLTASAV IRKAQVSELE RLMDSKIFNP EEREGVEKFA TTVKEGPILY NPSPSCSLEL KEKLETSHLK FDA SLRRLQ ELGKDSFLNM DNKFETYQKR LSIDYREILH CLDNLGLICA HLAAEVCLEK ISDTKEESET YKECSMVCKE FLED ILSTI GVYLPQDDKS LVDLQQNHLS AVISGHVSPK LKELFHLLDS FRGDKQKQCL ILVERIITAK VIERFVKKEA SLAYL NVLY LTENNPSTNV SAQKMQIEIP DLFQHGKVNL LFITDVVEEG FQVPDCSCMV CFDLPKTMCS YSQSQKHAKQ SNSKSI MFL ERGNPKQRDH LHDLMRREVL IQDPEAPNLK SCPPPVKNGH GVKEIGSMVI PDSNITVSEE AASTQTMSDP PSRNEQL PP CKKLRLDNNL LQSNGKEKVA SSKSKSSSSA AGSKKRKELH GTTCANALSG TWGENIDGAT FQAYKFDFCC NISGEVYS S FSLLLESTLA EDVGKVEMDL YLVRKLVKAS VSPCGQIRLS QEELVKAKYF QQFFFNGMFG KLFVGSKSQG TKREFLLQT DTSSLWHPAF MFLLLPVETN DLASSATIDW SAINSCASIV EFLKKNSLLD LRDSDGNQCN TSSGQEVLLD DKMEETNLIH FANASSDKN SLEELVVIAI HTGRIYSIVE AVSDSSAMSP FEVDASSGYA TYAEYFNKKY GIVLAHPNQP LMKLKQSHHA H NLLVDFNE EMVVKTEPKA GNVRKRKPNI HAHLPPELLA RIDVPRAVLK SIYLLPSVMH RLESLMLASQ LREEIDCSID NF SISSTSI LEAVTTLTCP ESFSMERLEL LGDSVLKYVA SCHLFLKYPD KDEGQLSRQR QSIISNSNLH RLTTSRKLQG YIR NGAFEP RRWTAPGQFS LFPVPCKCGI DTREVPLDPK FFTENMTIKI GKSCDMGHRW VVSKSVSDCA EALIGAYYVS GGLS ASLHM MKWLGIDVDF DPNLVVEAIN RVSLRCYIPK EDELIELERK IQHEFSAKFL LKEAITHSSL RESYSYERLE FLGDS VLDF LITRHLFNTY EQTGPGEMTD LRSACVNNEN FAQVAVKNNL HTHLQRCATV LETQINDYLM SFQKPDETGR SIPSIQ GPK ALGDVVESIA GALLIDTRLD LDQVWRVFEP LLSPLVTPDK LQLPPYRELN ELCDSLGYFF RVKCSNDGVK AQATIQL QL DDVLLTGDGS EQTNKLALGK AASHLLTQLE KRNISRKTSL GDNQSSMDVN LACNHSDRET LTSETTEIQS IVIPFIGP I NMKKGGPRGT LHEFCKKHLW PMPTFDTSEE KSRTPFEFID GGEKRTSFSS FTSTITLRIP NREAVMYAGE ARPDKKSSF DSAVVELLYE LERRKIVIIQ K |
-Macromolecule #2: TAS1a RNA forward strand (5'-phosphorylated)
| Macromolecule | Name: TAS1a RNA forward strand (5'-phosphorylated) / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:   Arabidopsis thaliana (thale cress) Arabidopsis thaliana (thale cress) |
| Molecular weight | Theoretical: 9.558729 KDa |
| Sequence | String: UACAAGCGAA UGAGUCAUUC AUCCUAAGUC |
-Macromolecule #3: TAS1a RNA reverse strand
| Macromolecule | Name: TAS1a RNA reverse strand / type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:   Arabidopsis thaliana (thale cress) Arabidopsis thaliana (thale cress) |
| Molecular weight | Theoretical: 10.267107 KDa |
| Sequence | String: GACUUAGGAU GAAUGACUCA UUCGCUUGUA GA |
-Macromolecule #4: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #5: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.8 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Software - Name: Gctf |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.73 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION / Number images used: 155929 |
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7vg3: |
 Movie
Movie Controller
Controller