[English] 日本語
 Yorodumi
Yorodumi- EMDB-31547: Cryo-EM structure of the human cholesterol transporter ABCG1 in c... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human cholesterol transporter ABCG1 in complex with cholesterol | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationABC-type sterol transporter activity / glycoprotein transport / cellular response to high density lipoprotein particle stimulus / intracellular cholesterol transport /  ABC transporters in lipid homeostasis / toxin transmembrane transporter activity / ABC transporters in lipid homeostasis / toxin transmembrane transporter activity /  floppase activity / positive regulation of cholesterol biosynthetic process / phospholipid homeostasis / phosphatidylcholine floppase activity ...ABC-type sterol transporter activity / glycoprotein transport / cellular response to high density lipoprotein particle stimulus / intracellular cholesterol transport / floppase activity / positive regulation of cholesterol biosynthetic process / phospholipid homeostasis / phosphatidylcholine floppase activity ...ABC-type sterol transporter activity / glycoprotein transport / cellular response to high density lipoprotein particle stimulus / intracellular cholesterol transport /  ABC transporters in lipid homeostasis / toxin transmembrane transporter activity / ABC transporters in lipid homeostasis / toxin transmembrane transporter activity /  floppase activity / positive regulation of cholesterol biosynthetic process / phospholipid homeostasis / phosphatidylcholine floppase activity / high-density lipoprotein particle remodeling / phospholipid efflux / cholesterol transfer activity / floppase activity / positive regulation of cholesterol biosynthetic process / phospholipid homeostasis / phosphatidylcholine floppase activity / high-density lipoprotein particle remodeling / phospholipid efflux / cholesterol transfer activity /  reverse cholesterol transport / Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate / low-density lipoprotein particle remodeling / HDL remodeling / cholesterol efflux / reverse cholesterol transport / Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate / low-density lipoprotein particle remodeling / HDL remodeling / cholesterol efflux /  cholesterol binding / regulation of cholesterol metabolic process / positive regulation of amyloid-beta formation / response to lipid / negative regulation of cholesterol storage / amyloid precursor protein catabolic process / negative regulation of macrophage derived foam cell differentiation / positive regulation of cholesterol efflux / NR1H3 & NR1H2 regulate gene expression linked to cholesterol transport and efflux / cholesterol metabolic process / cholesterol homeostasis / response to organic substance / cholesterol binding / regulation of cholesterol metabolic process / positive regulation of amyloid-beta formation / response to lipid / negative regulation of cholesterol storage / amyloid precursor protein catabolic process / negative regulation of macrophage derived foam cell differentiation / positive regulation of cholesterol efflux / NR1H3 & NR1H2 regulate gene expression linked to cholesterol transport and efflux / cholesterol metabolic process / cholesterol homeostasis / response to organic substance /  ADP binding / positive regulation of protein secretion / ADP binding / positive regulation of protein secretion /  phospholipid binding / transmembrane transport / recycling endosome / phospholipid binding / transmembrane transport / recycling endosome /  endosome / protein heterodimerization activity / external side of plasma membrane / endosome / protein heterodimerization activity / external side of plasma membrane /  Golgi membrane / endoplasmic reticulum membrane / Golgi membrane / endoplasmic reticulum membrane /  Golgi apparatus / Golgi apparatus /  ATP hydrolysis activity / protein homodimerization activity / ATP hydrolysis activity / protein homodimerization activity /  mitochondrion / mitochondrion /  ATP binding / ATP binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.26 Å cryo EM / Resolution: 3.26 Å | |||||||||
 Authors Authors | Xu D / Li YY / Yang FR / Sun CR / Pan JH / Wang L / Chen ZP / Fang SC / Yao XB / Hou WT ...Xu D / Li YY / Yang FR / Sun CR / Pan JH / Wang L / Chen ZP / Fang SC / Yao XB / Hou WT / Zhou CZ / Chen Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2022 Journal: Cell Rep / Year: 2022Title: Structure and transport mechanism of the human cholesterol transporter ABCG1. Authors: Da Xu / Yanyan Li / Fengrui Yang / Cai-Rong Sun / Jinheng Pan / Liang Wang / Zhi-Peng Chen / Shu-Cheng Fang / Xuebiao Yao / Wen-Tao Hou / Cong-Zhao Zhou / Yuxing Chen /  Abstract: The reverse cholesterol transport pathway is responsible for the maintenance of human cholesterol homeostasis, an imbalance of which usually leads to atherosclerosis. As a key component of this ...The reverse cholesterol transport pathway is responsible for the maintenance of human cholesterol homeostasis, an imbalance of which usually leads to atherosclerosis. As a key component of this pathway, the ATP-binding cassette transporter ABCG1 forwards cellular cholesterol to the extracellular acceptor nascent high-density lipoprotein (HDL). Here, we report a 3.26-Å cryo-electron microscopy structure of cholesterol-bound ABCG1 in an inward-facing conformation, which represents a turnover condition upon ATP binding. Structural analyses combined with functional assays reveals that a cluster of conserved hydrophobic residues, in addition to two sphingomyelins, constitute a well-defined cholesterol-binding cavity. The exit of this cavity is closed by three pairs of conserved Phe residues, which constitute a hydrophobic path for the release of cholesterol in an acceptor concentration-dependent manner. Overall, we propose an ABCG1-driven cholesterol transport cycle initiated by sphingomyelin-assisted cholesterol recruitment and accomplished by the release of cholesterol to HDL. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31547.map.gz emd_31547.map.gz | 49.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31547-v30.xml emd-31547-v30.xml emd-31547.xml emd-31547.xml | 12.7 KB 12.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31547.png emd_31547.png | 34.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31547 http://ftp.pdbj.org/pub/emdb/structures/EMD-31547 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31547 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31547 | HTTPS FTP |
-Related structure data
| Related structure data |  7fdvMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31547.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31547.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.01 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : The complex of human ABCG1 with cholesterol and ATP
| Entire | Name: The complex of human ABCG1 with cholesterol and ATP |
|---|---|
| Components |
|
-Supramolecule #1: The complex of human ABCG1 with cholesterol and ATP
| Supramolecule | Name: The complex of human ABCG1 with cholesterol and ATP / type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 152 KDa |
-Macromolecule #1: ATP-binding cassette sub-family G member 1
| Macromolecule | Name: ATP-binding cassette sub-family G member 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO EC number: Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 75.670844 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MACLMAAFSV GTAMNASSYS AEMTEPKSVC VSVDEVVSSN MEATETDLLN GHLKKVDNNL TEAQRFSSLP RRAAVNIEFR DLSYSVPEG PWWRKKGYKT LLKGISGKFN SGELVAIMGP SGAGKSTLMN ILAGYRETGM KGAVLINGLP RDLRCFRKVS C YIMQDDML ...String: MACLMAAFSV GTAMNASSYS AEMTEPKSVC VSVDEVVSSN MEATETDLLN GHLKKVDNNL TEAQRFSSLP RRAAVNIEFR DLSYSVPEG PWWRKKGYKT LLKGISGKFN SGELVAIMGP SGAGKSTLMN ILAGYRETGM KGAVLINGLP RDLRCFRKVS C YIMQDDML LPHLTVQEAM MVSAHLKLQE KDEGRREMVK EILTALGLLS CANTRTGSLS GGQRKRLAIA LELVNNPPVM FF DQPTSGL DSASCFQVVS LMKGLAQGGR SIICTIHQPS AKLFELFDQL YVLSQGQCVY RGKVCNLVPY LRDLGLNCPT YHN PADFVM EVASGEYGDQ NSRLVRAVRE GMCDSDHKRD LGGDAEVNPF LWHRPSEEVK QTKRLKGLRK DSSSMEGCHS FSAS CLTQF CILFKRTFLS IMRDSVLTHL RITSHIGIGL LIGLLYLGIG NEAKKVLSNS GFLFFSMLFL MFAALMPTVL TFPLE MGVF LREHLNYWYS LKAYYLAKTM ADVPFQIMFP VAYCSIVYWM TSQPSDAVRF VLFAALGTMT SLVAQSLGLL IGAAST SLQ VATFVGPVTA IPVLLFSGFF VSFDTIPTYL QWMSYISYVR YGFEGVILSI YGLDREDLHC DIDETCHFQK SEAILRE LD VENAKLYLDF IVLGIFFISL RLIAYFVLRY KIRAER |
-Macromolecule #2: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #3: [(E,2S,3R)-2-(hexadecanoylamino)-3-oxidanyl-octadec-4-enyl] 2-(tr...
| Macromolecule | Name: [(E,2S,3R)-2-(hexadecanoylamino)-3-oxidanyl-octadec-4-enyl] 2-(trimethylazaniumyl)ethyl phosphate type: ligand / ID: 3 / Number of copies: 2 / Formula: HWP |
|---|---|
| Molecular weight | Theoretical: 703.028 Da |
| Chemical component information | 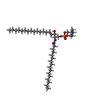 ChemComp-HWP: |
-Macromolecule #4: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 4 / Number of copies: 2 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 57.6 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Initial angle assignment | Type: PROJECTION MATCHING |
|---|---|
| Final angle assignment | Type: PROJECTION MATCHING |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.26 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 151593 |
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL / Overall B value: 142 |
|---|---|
| Output model |  PDB-7fdv: |
 Movie
Movie Controller
Controller











