[English] 日本語
 Yorodumi
Yorodumi- EMDB-29768: mRNA decoding in human is kinetically and structurally distinct f... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | mRNA decoding in human is kinetically and structurally distinct from bacteria (40S Focus refined map) | |||||||||
 Map data Map data | Unsharpened refine3D map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
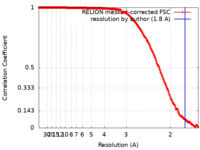
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 1.8 Å cryo EM / Resolution: 1.8 Å | |||||||||
 Authors Authors | Holm M / Natchiar KS / Rundlet EJ / Myasnikov AG / Altman RB / Blanchard SC | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: mRNA decoding in human is kinetically and structurally distinct from bacteria. Authors: Mikael Holm / S Kundhavai Natchiar / Emily J Rundlet / Alexander G Myasnikov / Zoe L Watson / Roger B Altman / Hao-Yuan Wang / Jack Taunton / Scott C Blanchard /   Abstract: In all species, ribosomes synthesize proteins by faithfully decoding messenger RNA (mRNA) nucleotide sequences using aminoacyl-tRNA substrates. Current knowledge of the decoding mechanism derives ...In all species, ribosomes synthesize proteins by faithfully decoding messenger RNA (mRNA) nucleotide sequences using aminoacyl-tRNA substrates. Current knowledge of the decoding mechanism derives principally from studies on bacterial systems. Although key features are conserved across evolution, eukaryotes achieve higher-fidelity mRNA decoding than bacteria. In human, changes in decoding fidelity are linked to ageing and disease and represent a potential point of therapeutic intervention in both viral and cancer treatment. Here we combine single-molecule imaging and cryogenic electron microscopy methods to examine the molecular basis of human ribosome fidelity to reveal that the decoding mechanism is both kinetically and structurally distinct from that of bacteria. Although decoding is globally analogous in both species, the reaction coordinate of aminoacyl-tRNA movement is altered on the human ribosome and the process is an order of magnitude slower. These distinctions arise from eukaryote-specific structural elements in the human ribosome and in the elongation factor eukaryotic elongation factor 1A (eEF1A) that together coordinate faithful tRNA incorporation at each mRNA codon. The distinct nature and timing of conformational changes within the ribosome and eEF1A rationalize how increased decoding fidelity is achieved and potentially regulated in eukaryotic species. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29768.map.gz emd_29768.map.gz | 807.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29768-v30.xml emd-29768-v30.xml emd-29768.xml emd-29768.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29768_fsc.xml emd_29768_fsc.xml | 22.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_29768.png emd_29768.png | 157.8 KB | ||
| Masks |  emd_29768_msk_1.map emd_29768_msk_1.map | 1000 MB |  Mask map Mask map | |
| Others |  emd_29768_additional_1.map.gz emd_29768_additional_1.map.gz emd_29768_additional_2.map.gz emd_29768_additional_2.map.gz emd_29768_half_map_1.map.gz emd_29768_half_map_1.map.gz emd_29768_half_map_2.map.gz emd_29768_half_map_2.map.gz | 932 MB 75.6 MB 810.3 MB 810.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29768 http://ftp.pdbj.org/pub/emdb/structures/EMD-29768 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29768 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29768 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29768.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29768.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened refine3D map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.826 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_29768_msk_1.map emd_29768_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
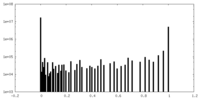
| Density Histograms |
-Additional map: Post-processed map
| File | emd_29768_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post-processed map | ||||||||||||
| Projections & Slices |
| ||||||||||||
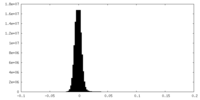
| Density Histograms |
-Additional map: Post-processed masked map
| File | emd_29768_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post-processed masked map | ||||||||||||
| Projections & Slices |
| ||||||||||||
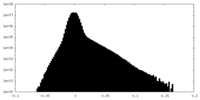
| Density Histograms |
-Half map: Half map 1
| File | emd_29768_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_29768_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
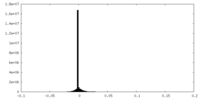
| Density Histograms |
- Sample components
Sample components
-Entire : Human ribosome
| Entire | Name: Human ribosome |
|---|---|
| Components |
|
-Supramolecule #1: Human ribosome
| Supramolecule | Name: Human ribosome / type: complex / ID: 1 / Chimera: Yes / Parent: 0 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 7 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: -1.5 µm / Nominal defocus min: -0.5 µm Bright-field microscopy / Nominal defocus max: -1.5 µm / Nominal defocus min: -0.5 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 79.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: OTHER |
|---|
 Movie
Movie Controller
Controller

















 Z
Z Y
Y X
X