[English] 日本語
 Yorodumi
Yorodumi- EMDB-29658: Mutant bacteriophage T4 gp41 helicase hexamer bound with single s... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mutant bacteriophage T4 gp41 helicase hexamer bound with single strand DNA and ATPgammaS in the stalled primosome | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  phage / phage /  complex / complex /  helicase / helicase /  REPLICATION REPLICATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationviral DNA genome replication /  DNA helicase activity / DNA helicase activity /  Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement /  DNA replication / DNA replication /  hydrolase activity / hydrolase activity /  DNA binding / DNA binding /  ATP binding ATP bindingSimilarity search - Function | |||||||||
| Biological species |   Escherichia phage T4 (virus) Escherichia phage T4 (virus) | |||||||||
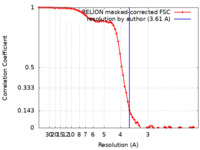
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.61 Å cryo EM / Resolution: 3.61 Å | |||||||||
 Authors Authors | Feng X / Li H | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation | Journal: bioRxiv / Year: 2023 Title: Structural basis of the T4 bacteriophage primosome assembly and primer synthesis. Authors: Xiang Feng / Michelle M Spiering / Ruda de Luna Almeida Santos / Stephen J Benkovic / Huilin Li /  Abstract: The T4 bacteriophage gp41 helicase and gp61 primase assemble into a primosome complex to couple DNA unwinding with RNA primer synthesis for DNA replication. How a primosome is assembled and how the ...The T4 bacteriophage gp41 helicase and gp61 primase assemble into a primosome complex to couple DNA unwinding with RNA primer synthesis for DNA replication. How a primosome is assembled and how the length of the RNA primer is defined in the T4 bacteriophage, or in any model system, are unclear. Here we report a series of cryo-EM structures of T4 primosome assembly intermediates at resolutions up to 2.7 Å. We show that the gp41 helicase is an open spiral in the absence of ssDNA, and ssDNA binding triggers a large-scale scissor-like conformational change that drives the open spiral to a closed ring that activates the helicase. We found that the activation of the gp41 helicase exposes a cryptic hydrophobic primase-binding surface allowing for the recruitment of the gp61 primase. The primase binds the gp41 helicase in a bipartite mode in which the N-terminal Zn-binding domain (ZBD) and the C-terminal RNA polymerase domain (RPD) each contain a helicase-interacting motif (HIM1 and HIM2, respectively) that bind to separate gp41 N-terminal hairpin dimers, leading to the assembly of one primase on the helicase hexamer. Based on two observed primosome conformations - one in a DNA-scanning mode and the other in a post RNA primer-synthesis mode - we suggest that the linker loop between the gp61 ZBD and RPD contributes to the T4 pentaribonucleotide primer. Our study reveals T4 primosome assembly process and sheds light on RNA primer synthesis mechanism. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29658.map.gz emd_29658.map.gz | 28.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29658-v30.xml emd-29658-v30.xml emd-29658.xml emd-29658.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29658_fsc.xml emd_29658_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_29658.png emd_29658.png | 122.8 KB | ||
| Others |  emd_29658_half_map_1.map.gz emd_29658_half_map_1.map.gz emd_29658_half_map_2.map.gz emd_29658_half_map_2.map.gz | 23.4 MB 23.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29658 http://ftp.pdbj.org/pub/emdb/structures/EMD-29658 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29658 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29658 | HTTPS FTP |
-Related structure data
| Related structure data |  8g0zMC  8dtpC  8dueC  8duoC  8dvfC  8dviC  8dw6C  8dwjC  8gaoC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29658.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29658.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.029 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_29658_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
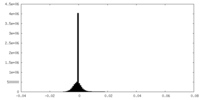
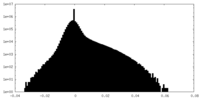
| Density Histograms |
-Half map: #1
| File | emd_29658_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
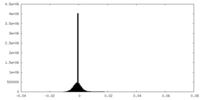
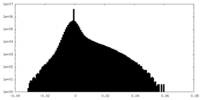
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of mutated T4 bacteriophage helicase gp41 with ssDNA/RNA ...
| Entire | Name: Complex of mutated T4 bacteriophage helicase gp41 with ssDNA/RNA hybrid |
|---|---|
| Components |
|
-Supramolecule #1: Complex of mutated T4 bacteriophage helicase gp41 with ssDNA/RNA ...
| Supramolecule | Name: Complex of mutated T4 bacteriophage helicase gp41 with ssDNA/RNA hybrid type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Escherichia phage T4 (virus) Escherichia phage T4 (virus) |
-Macromolecule #1: DnaB-like replicative helicase
| Macromolecule | Name: DnaB-like replicative helicase / type: protein_or_peptide / ID: 1 / Details: mutant Bacteriophage T4 / Number of copies: 6 / Enantiomer: LEVO EC number:  Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:   Escherichia phage T4 (virus) Escherichia phage T4 (virus) |
| Molecular weight | Theoretical: 48.796711 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: MVEIILSHLI FDQAYFSKVW PYMDSEYFES GPAKNTFKLI KSHVNEYHSV PSINALNVAL ENSSFTETEY SGVKTLISKL ADSPEDHSW LVKETEKYVQ QRAMFNATSK IIEIQTNAEL PPEKRNKKMP DVGAIPDIMR QALSISFDSY VGHDWMDDYE A RWLSYMNK ...String: MVEIILSHLI FDQAYFSKVW PYMDSEYFES GPAKNTFKLI KSHVNEYHSV PSINALNVAL ENSSFTETEY SGVKTLISKL ADSPEDHSW LVKETEKYVQ QRAMFNATSK IIEIQTNAEL PPEKRNKKMP DVGAIPDIMR QALSISFDSY VGHDWMDDYE A RWLSYMNK ARKVPFKLRI LNKITKGGAE TGTLNVLMAG VNVGKSLGLC SLAADYLQLG HNVLYISMQM AEEVCAKRID AN MLDVSLD DIDDGHISYA EYKGKMEKWR EKSTLGRLIV KQYPTGGADA NTFRSLLNEL KLKKNFVPTI IIVDYLGICK SCR IRVYSE NSYTTVKAIA EELRALAVET ETVLWTAAQV GKQAWDSSDV NMSDIAESAG LPATADFMLA VIETEELAAA EQQL IKQIK SRYGDKNKWN KFLMGVQKGN QKWVEIE UniProtKB: DnaB-like replicative helicase |
-Macromolecule #2: DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3')
| Macromolecule | Name: DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3') / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Escherichia phage T4 (virus) Escherichia phage T4 (virus) |
| Molecular weight | Theoretical: 3.605356 KDa |
| Sequence | String: (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT) |
-Macromolecule #3: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER / type: ligand / ID: 3 / Number of copies: 5 / Formula: AGS |
|---|---|
| Molecular weight | Theoretical: 523.247 Da |
| Chemical component information |  ChemComp-AGS: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 5 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 Component:
Details: 20 mM HEPES pH 7.8, 100 mM NaCl, 10 mM MgCl2 and 2 mM DTT | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 66.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-8g0z: |
 Movie
Movie Controller
Controller















 Z
Z Y
Y X
X