[English] 日本語
 Yorodumi
Yorodumi- EMDB-29406: Pseudomonas phage E217 5-fold vertex (capsid and decorating proteins) -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Pseudomonas phage E217 5-fold vertex (capsid and decorating proteins) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Pseudomonas / Pseudomonas /  phage / E217 / phage / E217 /  VIRUS / VIRUS /  capsid / decorating proteins capsid / decorating proteins | |||||||||
| Function / homology | Protein of unknown function DUF2184 / Uncharacterized protein conserved in bacteria (DUF2184) / Capsid and scaffold protein /  Virion protein Virion protein Function and homology information Function and homology information | |||||||||
| Biological species |  Pseudomonas phage vB_PaeM_E217 (virus) Pseudomonas phage vB_PaeM_E217 (virus) | |||||||||
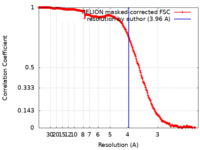
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.96 Å cryo EM / Resolution: 3.96 Å | |||||||||
 Authors Authors | Li F / Cingolani G / Hou C | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: High-resolution cryo-EM structure of the Pseudomonas bacteriophage E217. Authors: Fenglin Li / Chun-Feng David Hou / Ravi K Lokareddy / Ruoyu Yang / Francesca Forti / Federica Briani / Gino Cingolani /   Abstract: E217 is a Pseudomonas phage used in an experimental cocktail to eradicate cystic fibrosis-associated Pseudomonas aeruginosa. Here, we describe the structure of the whole E217 virion before and after ...E217 is a Pseudomonas phage used in an experimental cocktail to eradicate cystic fibrosis-associated Pseudomonas aeruginosa. Here, we describe the structure of the whole E217 virion before and after DNA ejection at 3.1 Å and 4.5 Å resolution, respectively, determined using cryogenic electron microscopy (cryo-EM). We identify and build de novo structures for 19 unique E217 gene products, resolve the tail genome-ejection machine in both extended and contracted states, and decipher the complete architecture of the baseplate formed by 66 polypeptide chains. We also determine that E217 recognizes the host O-antigen as a receptor, and we resolve the N-terminal portion of the O-antigen-binding tail fiber. We propose that E217 design principles presented in this paper are conserved across PB1-like Myoviridae phages of the Pbunavirus genus that encode a ~1.4 MDa baseplate, dramatically smaller than the coliphage T4. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29406.map.gz emd_29406.map.gz | 407.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29406-v30.xml emd-29406-v30.xml emd-29406.xml emd-29406.xml | 14.6 KB 14.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29406_fsc.xml emd_29406_fsc.xml | 18 KB | Display |  FSC data file FSC data file |
| Images |  emd_29406.png emd_29406.png | 956.2 KB | ||
| Others |  emd_29406_half_map_1.map.gz emd_29406_half_map_1.map.gz emd_29406_half_map_2.map.gz emd_29406_half_map_2.map.gz | 411.7 MB 411.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29406 http://ftp.pdbj.org/pub/emdb/structures/EMD-29406 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29406 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29406 | HTTPS FTP |
-Related structure data
| Related structure data |  8frsMC  8envC  8eonC  8fuvC  8fvgC  8fvhC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29406.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29406.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.12 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_29406_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_29406_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Pseudomonas phage vB_PaeM_E217
| Entire | Name:  Pseudomonas phage vB_PaeM_E217 (virus) Pseudomonas phage vB_PaeM_E217 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Pseudomonas phage vB_PaeM_E217
| Supramolecule | Name: Pseudomonas phage vB_PaeM_E217 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2034346 / Sci species name: Pseudomonas phage vB_PaeM_E217 / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: Yes / Virus empty: Yes |
|---|
-Macromolecule #1: Major structural protein
| Macromolecule | Name: Major structural protein / type: protein_or_peptide / ID: 1 / Number of copies: 11 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage vB_PaeM_E217 (virus) Pseudomonas phage vB_PaeM_E217 (virus) |
| Molecular weight | Theoretical: 34.735047 KDa |
| Sequence | String: NFTAPVTTPS IPTPIQFLQT WLPGFVKVMT AARKIDEIIG IDTVGSWEDQ EIVQGIVEPA GTAVEYGDHT NIPLTSWNAN FERRTIVRG ELGMMVGTLE EGRASAIRLN SAETKRQQAA IGLETFRNAI GFYGWQSGLG NRTYGFLNDP NLPAFQTPPS Q GWSTADWA ...String: NFTAPVTTPS IPTPIQFLQT WLPGFVKVMT AARKIDEIIG IDTVGSWEDQ EIVQGIVEPA GTAVEYGDHT NIPLTSWNAN FERRTIVRG ELGMMVGTLE EGRASAIRLN SAETKRQQAA IGLETFRNAI GFYGWQSGLG NRTYGFLNDP NLPAFQTPPS Q GWSTADWA GIIGDIREAV RQLRIQSQDQ IDPKAEKITL ALATSKVDYL SVTTPYGISV SDWIEQTYPK MRIVSAPELS GV QMKNQEP EDALVLFVED VNAAVDGSTD GGSVFSQLVQ SKFITLGVEK RAKSYVEDFS NGTAGALCKR PWAVVRYLGI UniProtKB: Capsid and scaffold protein |
-Macromolecule #2: Structural protein gp24
| Macromolecule | Name: Structural protein gp24 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage vB_PaeM_E217 (virus) Pseudomonas phage vB_PaeM_E217 (virus) |
| Molecular weight | Theoretical: 21.610312 KDa |
| Sequence | String: MFQKQVYRQY TPGFPGDLIE DGPKRARPGR IMSLSAVNPA ATATGPNRAS RAFGYAGDVS ALGEGQPKTI AARASEVVIG GANFFGVLG HPKHYALFGS AGDSLAPSYD LPDGAEGEFF DMATGLVVEI FNGAAAALDL DYGDLVAYVP NNLATADDAL G LPAGALVG ...String: MFQKQVYRQY TPGFPGDLIE DGPKRARPGR IMSLSAVNPA ATATGPNRAS RAFGYAGDVS ALGEGQPKTI AARASEVVIG GANFFGVLG HPKHYALFGS AGDSLAPSYD LPDGAEGEFF DMATGLVVEI FNGAAAALDL DYGDLVAYVP NNLATADDAL G LPAGALVG FKTGSMPTGL VQIPNARIVN AISLPAQSAG NLVAGVTIVQ LTQ UniProtKB:  Virion protein Virion protein |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: OTHER / Imaging mode: OTHER / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 81000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z
Z Y
Y X
X