[English] 日本語
 Yorodumi
Yorodumi- EMDB-29302: CryoEM structure of Go-coupled NTSR1 with a biased allosteric mod... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
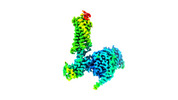
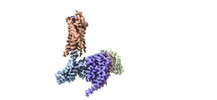
| Title | CryoEM structure of Go-coupled NTSR1 with a biased allosteric modulator | |||||||||
 Map data Map data | Go-coupled NTSR1 with a biased allosteric modulator | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to antipsychotic drug / Peptide ligand-binding receptors /  neuropeptide receptor binding / G protein-coupled neurotensin receptor activity / inositol phosphate catabolic process / positive regulation of inhibitory postsynaptic potential / symmetric synapse / response to mineralocorticoid / D-aspartate import across plasma membrane / positive regulation of gamma-aminobutyric acid secretion ...response to antipsychotic drug / Peptide ligand-binding receptors / neuropeptide receptor binding / G protein-coupled neurotensin receptor activity / inositol phosphate catabolic process / positive regulation of inhibitory postsynaptic potential / symmetric synapse / response to mineralocorticoid / D-aspartate import across plasma membrane / positive regulation of gamma-aminobutyric acid secretion ...response to antipsychotic drug / Peptide ligand-binding receptors /  neuropeptide receptor binding / G protein-coupled neurotensin receptor activity / inositol phosphate catabolic process / positive regulation of inhibitory postsynaptic potential / symmetric synapse / response to mineralocorticoid / D-aspartate import across plasma membrane / positive regulation of gamma-aminobutyric acid secretion / L-glutamate import across plasma membrane / positive regulation of arachidonic acid secretion / neuron spine / regulation of respiratory gaseous exchange / neuropeptide hormone activity / digestive tract development / negative regulation of systemic arterial blood pressure / hyperosmotic response / negative regulation of release of sequestered calcium ion into cytosol / G alpha (q) signalling events / positive regulation of glutamate secretion / positive regulation of inositol phosphate biosynthetic process / neuropeptide receptor binding / G protein-coupled neurotensin receptor activity / inositol phosphate catabolic process / positive regulation of inhibitory postsynaptic potential / symmetric synapse / response to mineralocorticoid / D-aspartate import across plasma membrane / positive regulation of gamma-aminobutyric acid secretion / L-glutamate import across plasma membrane / positive regulation of arachidonic acid secretion / neuron spine / regulation of respiratory gaseous exchange / neuropeptide hormone activity / digestive tract development / negative regulation of systemic arterial blood pressure / hyperosmotic response / negative regulation of release of sequestered calcium ion into cytosol / G alpha (q) signalling events / positive regulation of glutamate secretion / positive regulation of inositol phosphate biosynthetic process /  temperature homeostasis / response to corticosterone / response to lipid / temperature homeostasis / response to corticosterone / response to lipid /  regulation of membrane depolarization / cellular response to lithium ion / detection of temperature stimulus involved in sensory perception of pain / neuropeptide signaling pathway / response to axon injury / axon terminus / regulation of membrane depolarization / cellular response to lithium ion / detection of temperature stimulus involved in sensory perception of pain / neuropeptide signaling pathway / response to axon injury / axon terminus /  transport vesicle / response to amphetamine / blood vessel diameter maintenance / cellular response to dexamethasone stimulus / adult locomotory behavior / cellular response to nerve growth factor stimulus / positive regulation of release of sequestered calcium ion into cytosol / dendritic shaft / response to cocaine / liver development / transport vesicle / response to amphetamine / blood vessel diameter maintenance / cellular response to dexamethasone stimulus / adult locomotory behavior / cellular response to nerve growth factor stimulus / positive regulation of release of sequestered calcium ion into cytosol / dendritic shaft / response to cocaine / liver development /  learning / Olfactory Signaling Pathway / Activation of the phototransduction cascade / learning / Olfactory Signaling Pathway / Activation of the phototransduction cascade /  visual learning / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / visual learning / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 /  terminal bouton / cytoplasmic side of plasma membrane / ADP signalling through P2Y purinoceptor 12 / G beta:gamma signalling through BTK / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / photoreceptor disc membrane / Adrenaline,noradrenaline inhibits insulin secretion / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / cellular response to catecholamine stimulus / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / adenylate cyclase-activating dopamine receptor signaling pathway / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / cellular response to prostaglandin E stimulus / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / sensory perception of taste / GPER1 signaling / G-protein beta-subunit binding / terminal bouton / cytoplasmic side of plasma membrane / ADP signalling through P2Y purinoceptor 12 / G beta:gamma signalling through BTK / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / photoreceptor disc membrane / Adrenaline,noradrenaline inhibits insulin secretion / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / cellular response to catecholamine stimulus / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / adenylate cyclase-activating dopamine receptor signaling pathway / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / cellular response to prostaglandin E stimulus / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / sensory perception of taste / GPER1 signaling / G-protein beta-subunit binding /  heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade /  extracellular vesicle / G alpha (12/13) signalling events / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / response to estradiol / retina development in camera-type eye / extracellular vesicle / G alpha (12/13) signalling events / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / response to estradiol / retina development in camera-type eye /  GTPase binding / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (i) signalling events / fibroblast proliferation / GTPase binding / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (i) signalling events / fibroblast proliferation /  perikaryon / G alpha (s) signalling events / G alpha (q) signalling events / Ras protein signal transduction / cell population proliferation / perikaryon / G alpha (s) signalling events / G alpha (q) signalling events / Ras protein signal transduction / cell population proliferation /  dendritic spine / Extra-nuclear estrogen signaling / dendritic spine / Extra-nuclear estrogen signaling /  receptor ligand activity / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / positive regulation of apoptotic process receptor ligand activity / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / positive regulation of apoptotic processSimilarity search - Function | |||||||||
| Biological species |   Rattus norvegicus (Norway rat) / Rattus norvegicus (Norway rat) /   Homo sapiens (human) / Homo sapiens (human) /   Escherichia coli (E. coli) / Escherichia coli (E. coli) /   Lama glama (llama) Lama glama (llama) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.89 Å cryo EM / Resolution: 2.89 Å | |||||||||
 Authors Authors | Krumm BE / DiBerto JF / Olsen RHJ / Kang H / Slocum ST / Zhang S / Strachan RT / Fay JF / Roth BL | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Biochemistry / Year: 2023 Journal: Biochemistry / Year: 2023Title: Neurotensin Receptor Allosterism Revealed in Complex with a Biased Allosteric Modulator. Authors: Brian E Krumm / Jeffrey F DiBerto / Reid H J Olsen / Hye Jin Kang / Samuel T Slocum / Shicheng Zhang / Ryan T Strachan / Xi-Ping Huang / Lauren M Slosky / Anthony B Pinkerton / Lawrence S ...Authors: Brian E Krumm / Jeffrey F DiBerto / Reid H J Olsen / Hye Jin Kang / Samuel T Slocum / Shicheng Zhang / Ryan T Strachan / Xi-Ping Huang / Lauren M Slosky / Anthony B Pinkerton / Lawrence S Barak / Marc G Caron / Terry Kenakin / Jonathan F Fay / Bryan L Roth /  Abstract: The NTSR1 neurotensin receptor (NTSR1) is a G protein-coupled receptor (GPCR) found in the brain and peripheral tissues with neurotensin (NTS) being its endogenous peptide ligand. In the brain, NTS ...The NTSR1 neurotensin receptor (NTSR1) is a G protein-coupled receptor (GPCR) found in the brain and peripheral tissues with neurotensin (NTS) being its endogenous peptide ligand. In the brain, NTS modulates dopamine neuronal activity, induces opioid-independent analgesia, and regulates food intake. Recent studies indicate that biasing NTSR1 toward β-arrestin signaling can attenuate the actions of psychostimulants and other drugs of abuse. Here, we provide the cryoEM structures of NTSR1 ternary complexes with heterotrimeric Gq and GoA with and without the brain-penetrant small-molecule SBI-553. In functional studies, we discovered that SBI-553 displays complex allosteric actions exemplified by negative allosteric modulation for G proteins that are Gα subunit selective and positive allosteric modulation and agonism for β-arrestin translocation at NTSR1. Detailed structural analysis of the allosteric binding site illuminated the structural determinants for biased allosteric modulation of SBI-553 on NTSR1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29302.map.gz emd_29302.map.gz | 55.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29302-v30.xml emd-29302-v30.xml emd-29302.xml emd-29302.xml | 20.7 KB 20.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29302.png emd_29302.png | 69.3 KB | ||
| Others |  emd_29302_half_map_1.map.gz emd_29302_half_map_1.map.gz emd_29302_half_map_2.map.gz emd_29302_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29302 http://ftp.pdbj.org/pub/emdb/structures/EMD-29302 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29302 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29302 | HTTPS FTP |
-Related structure data
| Related structure data |  8fn0MC  8fmzC  8fn1C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29302.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29302.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Go-coupled NTSR1 with a biased allosteric modulator | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.88 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map 1
| File | emd_29302_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map 2
| File | emd_29302_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of NTSR1 MiniGo heterotrimer with scFv16
| Entire | Name: Ternary complex of NTSR1 MiniGo heterotrimer with scFv16 |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of NTSR1 MiniGo heterotrimer with scFv16
| Supramolecule | Name: Ternary complex of NTSR1 MiniGo heterotrimer with scFv16 type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:   Rattus norvegicus (Norway rat) Rattus norvegicus (Norway rat) |
| Molecular weight | Theoretical: 147 KDa |
-Macromolecule #1: Neurotensin receptor type 1
| Macromolecule | Name: Neurotensin receptor type 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Rattus norvegicus (Norway rat) Rattus norvegicus (Norway rat) |
| Molecular weight | Theoretical: 45.939211 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MHHHHHHHHH HSDLEVLFQG PLGSGAPTSE SDTAGPNSDL DVNTDIYSKV LVTAIYLALF VVGTVGNSVT LFTLARKKSL QSLQSTVHY HLGSLALSDL LILLLAMPVE LYNFIWVHHP WAFGDAGCRG YYFLRDACTY ATALNVASLS VERYLAICHP F KAKTLMSR ...String: MHHHHHHHHH HSDLEVLFQG PLGSGAPTSE SDTAGPNSDL DVNTDIYSKV LVTAIYLALF VVGTVGNSVT LFTLARKKSL QSLQSTVHY HLGSLALSDL LILLLAMPVE LYNFIWVHHP WAFGDAGCRG YYFLRDACTY ATALNVASLS VERYLAICHP F KAKTLMSR SRTKKFISAI WLASALLAIP MLFTMGLQNR SADGTHPGGL VCTPIVDTAT VKVVIQVNTF MSFLFPMLVI SI LNTVIAN KLTVMVHQAA EQGRVCTVGT HNGLEHSTFN MTIEPGRVQA LRHGVLVLRA VVIAFVVCWL PYHVRRLMFC YIS DEQWTT FLFDFYHYFY MLTNALFYAS SAINPILYNL VSANFRQVFL STLACLCPGW RHRRKKRPTF SRKPNSMSSN HAFS TSATR ETLY |
-Macromolecule #2: Neurotensin/neuromedin N
| Macromolecule | Name: Neurotensin/neuromedin N / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Rattus norvegicus (Norway rat) Rattus norvegicus (Norway rat) |
| Molecular weight | Theoretical: 819.007 Da |
| Sequence | String: RRPYIL |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.418086 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR ...String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR FLDDNQIVTS SGDTTCALWD IETGQQTTTF TGHTGDVMSL SLAPDTRLFV SGACDASAKL WDVREGMCRQ TF TGHESDI NAICFFPNGN AFATGSDDAT CRLFDLRADQ ELMTYSHDNI ICGITSVSFS KSGRLLLAGY DDFNCNVWDA LKA DRAGVL AGHDNRVSCL GVTDDGMAVA TGSWDSFLKI WN |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L |
-Macromolecule #5: MiniGo
| Macromolecule | Name: MiniGo / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Molecular weight | Theoretical: 25.451166 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MGCTLSAEDK AAVERSKMIE KNLKEDGISA AKDVKLLLLG ADNSGKSTIV KQMKIIHGGS GGSGGTTGIV ETHFTFKNLH FRLFDVGGQ RSERKKWIHC FEDVTAIIFC VDLSDYNRMH ESLMLFDSIC NNKFFIDTSI ILFLNKKDLF GEKIKKSPLT I CFPEYTGP ...String: MGCTLSAEDK AAVERSKMIE KNLKEDGISA AKDVKLLLLG ADNSGKSTIV KQMKIIHGGS GGSGGTTGIV ETHFTFKNLH FRLFDVGGQ RSERKKWIHC FEDVTAIIFC VDLSDYNRMH ESLMLFDSIC NNKFFIDTSI ILFLNKKDLF GEKIKKSPLT I CFPEYTGP NTYEDAAAYI QAQFESKNRS PNKEIYCHMT CATDTNNAQV IFDAVTDIII ANNLRGCGLY |
-Macromolecule #6: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Lama glama (llama) Lama glama (llama) |
| Molecular weight | Theoretical: 28.668922 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KAAALEVLFQ GPHHHHHHHH |
-Macromolecule #7: 2-[{2-(1-fluorocyclopropyl)-4-[4-(2-methoxyphenyl)piperidin-1-yl]...
| Macromolecule | Name: 2-[{2-(1-fluorocyclopropyl)-4-[4-(2-methoxyphenyl)piperidin-1-yl]quinazolin-6-yl}(methyl)amino]ethan-1-ol type: ligand / ID: 7 / Number of copies: 1 / Formula: SRW |
|---|---|
| Molecular weight | Theoretical: 450.548 Da |
| Chemical component information |  ChemComp-SRW: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 20mM Hepes, 0.1M NaCl, 0.00075% LMNG, 0.00075% GDN |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.2 µm / Nominal defocus min: 0.1 µm Bright-field microscopy / Nominal defocus max: 3.2 µm / Nominal defocus min: 0.1 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 44.0 e/Å2 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.89 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 461511 |
 Movie
Movie Controller
Controller
























 Z
Z Y
Y X
X