[English] 日本語
 Yorodumi
Yorodumi- EMDB-29209: Structure of Bispecific CAP256V2LS-J3 Fab in complex with BG505 D... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 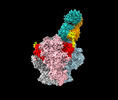 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
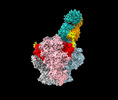
| Title | Structure of Bispecific CAP256V2LS-J3 Fab in complex with BG505 DS-SOSIP.664 | |||||||||
 Map data Map data | Sharpened map | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / virus-mediated perturbation of host defense response / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell /  viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope ...positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / virus-mediated perturbation of host defense response / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral envelope ...positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / virus-mediated perturbation of host defense response / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell /  viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / identical protein binding / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / identical protein binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |    Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /   Lama glama (llama) / Lama glama (llama) /   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.18 Å cryo EM / Resolution: 3.18 Å | |||||||||
 Authors Authors | Gorman J / Kwong PD | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: MAbs / Year: 2023 Journal: MAbs / Year: 2023Title: Bispecific antibody CAP256.J3LS targets V2-apex and CD4-binding sites with high breadth and potency. Authors: Baoshan Zhang / Jason Gorman / Young D Kwon / Amarendra Pegu / Cara W Chao / Tracy Liu / Mangaiarkarasi Asokan / Michael F Bender / Tatsiana Bylund / Leland Damron / Deepika Gollapudi / ...Authors: Baoshan Zhang / Jason Gorman / Young D Kwon / Amarendra Pegu / Cara W Chao / Tracy Liu / Mangaiarkarasi Asokan / Michael F Bender / Tatsiana Bylund / Leland Damron / Deepika Gollapudi / Paula Lei / Yile Li / Cuiping Liu / Mark K Louder / Krisha McKee / Adam S Olia / Reda Rawi / Arne Schön / Shuishu Wang / Eun Sung Yang / Yongping Yang / Kevin Carlton / Nicole A Doria-Rose / Lawrence Shapiro / Michael S Seaman / John R Mascola / Peter D Kwong /  Abstract: Antibody CAP256-VRC26.25 targets the second hypervariable region (V2) at the apex of the HIV envelope (Env) trimer with extraordinary neutralization potency, although less than optimal breadth. To ...Antibody CAP256-VRC26.25 targets the second hypervariable region (V2) at the apex of the HIV envelope (Env) trimer with extraordinary neutralization potency, although less than optimal breadth. To improve breadth, we linked the light chain of CAP256V2LS, an optimized version of CAP256-VRC26.25 currently under clinical evaluation, to the llama nanobody J3, which has broad CD4-binding site-directed neutralization. The J3-linked bispecific antibody exhibited improved breadth and potency over both J3 and CAP256V2LS, indicative of synergistic neutralization. The cryo-EM structure of the bispecific antibody in complex with a prefusion-closed Env trimer revealed simultaneous binding of J3 and CAP256V2LS. We further optimized the pharmacokinetics of the bispecific antibody by reducing the net positive charge of J3. The optimized bispecific antibody, which we named CAP256.J3LS, had a half-life similar to CAP256V2LS in human FcRn knock-in mice and exhibited suitable auto-reactivity, manufacturability, and biophysical risk. CAP256.J3LS neutralized over 97% of a multiclade 208-strain panel (geometric mean concentration for 80% inhibition (IC) 0.079 μg/ml) and 100% of a 100-virus clade C panel (geometric mean IC of 0.05 μg/ml), suggesting its anti-HIV utility especially in regions where clade C dominates. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29209.map.gz emd_29209.map.gz | 228.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29209-v30.xml emd-29209-v30.xml emd-29209.xml emd-29209.xml | 25.1 KB 25.1 KB | Display Display |  EMDB header EMDB header |
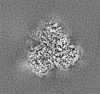
| Images |  emd_29209.png emd_29209.png | 72.9 KB | ||
| Masks |  emd_29209_msk_1.map emd_29209_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Others |  emd_29209_additional_1.map.gz emd_29209_additional_1.map.gz emd_29209_additional_2.map.gz emd_29209_additional_2.map.gz emd_29209_half_map_1.map.gz emd_29209_half_map_1.map.gz emd_29209_half_map_2.map.gz emd_29209_half_map_2.map.gz | 60.1 MB 21 MB 226.2 MB 226.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29209 http://ftp.pdbj.org/pub/emdb/structures/EMD-29209 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29209 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29209 | HTTPS FTP |
-Related structure data
| Related structure data |  8fisMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29209.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29209.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.083 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_29209_msk_1.map emd_29209_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened
| File | emd_29209_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: resolve density modified
| File | emd_29209_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | resolve density modified | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A
| File | emd_29209_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B
| File | emd_29209_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : CAP256V2LS-J3-3 Fab in complex with BG505 DS-SOSIP.664
| Entire | Name: CAP256V2LS-J3-3 Fab in complex with BG505 DS-SOSIP.664 |
|---|---|
| Components |
|
-Supramolecule #1: CAP256V2LS-J3-3 Fab in complex with BG505 DS-SOSIP.664
| Supramolecule | Name: CAP256V2LS-J3-3 Fab in complex with BG505 DS-SOSIP.664 type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:    Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Macromolecule #1: Envelope glycoprotein gp41
| Macromolecule | Name: Envelope glycoprotein gp41 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17.146482 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAVFLG FLGAAGSTMG AASMTLTVQA RNLLSGIVQQ QSNLLRAPEA QQHLLKLTVW GIKQLQARVL AVERYLRDQQ LLGIWGCSG KLICCTNVPW NSSWSNRNLS EIWDNMTWLQ WDKEISNYTQ IIYGLLEESQ NQQEKNEQDL LALD |
-Macromolecule #2: Envelope glycoprotein gp120
| Macromolecule | Name: Envelope glycoprotein gp120 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 54.086324 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETEKHNVWA THACVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHTDIIS LWDQSLKPC VKLTPLCVTL QCTNVTNNIT DDMRGELKNC SFNMTTELRD KKQKVYSLFY RLDVVQINEN QGNRSNNSNK E YRLINCNT ...String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETEKHNVWA THACVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHTDIIS LWDQSLKPC VKLTPLCVTL QCTNVTNNIT DDMRGELKNC SFNMTTELRD KKQKVYSLFY RLDVVQINEN QGNRSNNSNK E YRLINCNT SACTQACPKV SFEPIPIHYC APAGFAILKC KDKKFNGTGP CPSVSTVQCT HGIKPVVSTQ LLLNGSLAEE EV MIRSENI TNNAKNILVQ FNTPVQINCT RPNNNTRKSI RIGPGQAFYA TGDIIGDIRQ AHCNVSKATW NETLGKVVKQ LRK HFGNNT IIRFANSSGG DLEVTTHSFN CGGEFFYCNT SGLFNSTWIS NTSVQGSNST GSNDSITLPC RIKQIINMWQ RIGQ CMYAP PIQGVIRCVS NITGLILTRD GGSTNSTTET FRPGGGDMRD NWRSELYKYK VVKIEPLGVA PTRCKRRVVG RRRRR R |
-Macromolecule #3: J3-VRC26.25 Light
| Macromolecule | Name: J3-VRC26.25 Light / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Lama glama (llama) Lama glama (llama) |
| Molecular weight | Theoretical: 37.355211 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGGG LVQAGGFLEL SCELRGSIFN QYAMAWFRQA PGKEREFVAG MGAVPHYGEF VKGRFTISRD NAKSTVYLQM SSLEPEDTA IYFCARSKST YISYNSNGYD YWGQGTQVTV SSGGSGGGGS GGGGSGGQSV LTQPPSVSAA PGQKVTISCS G NTSNIGNN ...String: EVQLVESGGG LVQAGGFLEL SCELRGSIFN QYAMAWFRQA PGKEREFVAG MGAVPHYGEF VKGRFTISRD NAKSTVYLQM SSLEPEDTA IYFCARSKST YISYNSNGYD YWGQGTQVTV SSGGSGGGGS GGGGSGGQSV LTQPPSVSAA PGQKVTISCS G NTSNIGNN FVSWYQQRPG RAPQLLIYET DKRPSGIPDR FSASKSGTSG TLAITGLQTG DEADYYCATW AASLSSARVF GT GTQVIVL GQPKVNPTVT LFPPSSEELQ ANKATLVCLI SDFYPGAVTV AWKADSSPVK AGVETTTPSK QSNNKYAASS YLS LTPEQW KSHRSYSCQV THEGSTVEKT VAPTECS |
-Macromolecule #4: VRC26.25 Heavy
| Macromolecule | Name: VRC26.25 Heavy / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.863039 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVESGGG VVQPGTSLRL SCAASQFRFD GYGMHWVRQA PGKGLEWVAS ISHDGIKKYH AEKVWGRFTI SRDNSKNTLY LQMNSLRPE DTALYYCAKD LREDECEEWW SD(TYS)(TYS)DFGAQL PCAKSRGGLV GIADNWGQGT MVTVSSASTK GPS VFPLAP ...String: QVQLVESGGG VVQPGTSLRL SCAASQFRFD GYGMHWVRQA PGKGLEWVAS ISHDGIKKYH AEKVWGRFTI SRDNSKNTLY LQMNSLRPE DTALYYCAKD LREDECEEWW SD(TYS)(TYS)DFGAQL PCAKSRGGLV GIADNWGQGT MVTVSSASTK GPS VFPLAP SSKSTSGGTA ALGCLVKDYF PEPVTVSWNS GALTSGVHTF PAVLQSSGLY SLSSVVTVPS SSLGTQTYIC NVNH KPSNT KVDKKVEPKS |
-Macromolecule #10: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 10 / Number of copies: 25 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: PBS |
| Grid | Model: C-flat-1.2/1.3 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Average exposure time: 2.0 sec. / Average electron dose: 51.19 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Initial angle assignment | Type: NOT APPLICABLE |
|---|---|
| Final 3D classification | Software - Name: cryoSPARC (ver. 3) |
| Final angle assignment | Type: NOT APPLICABLE |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.18 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 3) / Number images used: 118472 |
 Movie
Movie Controller
Controller





 Z
Z Y
Y X
X