[English] 日本語
 Yorodumi
Yorodumi- EMDB-29051: Wildtype rabbit TRPV5 into nanodiscs in the presence of PI(4,5)P2... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Wildtype rabbit TRPV5 into nanodiscs in the presence of PI(4,5)P2 and ruthenium red | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  TRPV5 / TRPV5 /  TRP Channel / TRP Channel /  Ruthenium Red / PI(4 / 5)P2 / Ruthenium Red / PI(4 / 5)P2 /  MEMBRANE PROTEIN MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of urine volume / calcium ion import across plasma membrane / calcium ion homeostasis / calcium ion transmembrane transport /  calcium channel activity / calcium ion transport / protein homotetramerization / calcium channel activity / calcium ion transport / protein homotetramerization /  calmodulin binding / apical plasma membrane / identical protein binding ...regulation of urine volume / calcium ion import across plasma membrane / calcium ion homeostasis / calcium ion transmembrane transport / calmodulin binding / apical plasma membrane / identical protein binding ...regulation of urine volume / calcium ion import across plasma membrane / calcium ion homeostasis / calcium ion transmembrane transport /  calcium channel activity / calcium ion transport / protein homotetramerization / calcium channel activity / calcium ion transport / protein homotetramerization /  calmodulin binding / apical plasma membrane / identical protein binding / calmodulin binding / apical plasma membrane / identical protein binding /  metal ion binding / metal ion binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Oryctolagus cuniculus (rabbit) Oryctolagus cuniculus (rabbit) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.65 Å cryo EM / Resolution: 2.65 Å | |||||||||
 Authors Authors | De Jesus-Perez JJ / Fluck EC / Pumroy RA / Protopopova AD / Rocereta JA / Moiseenkova-Bell VY | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: EMBO Rep / Year: 2024 Journal: EMBO Rep / Year: 2024Title: Molecular details of ruthenium red pore block in TRPV channels. Authors: Ruth A Pumroy / José J De Jesús-Pérez / Anna D Protopopova / Julia A Rocereta / Edwin C Fluck / Tabea Fricke / Bo-Hyun Lee / Tibor Rohacs / Andreas Leffler / Vera Moiseenkova-Bell /    Abstract: Transient receptor potential vanilloid (TRPV) channels play a critical role in calcium homeostasis, pain sensation, immunological response, and cancer progression. TRPV channels are blocked by ...Transient receptor potential vanilloid (TRPV) channels play a critical role in calcium homeostasis, pain sensation, immunological response, and cancer progression. TRPV channels are blocked by ruthenium red (RR), a universal pore blocker for a wide array of cation channels. Here we use cryo-electron microscopy to reveal the molecular details of RR block in TRPV2 and TRPV5, members of the two TRPV subfamilies. In TRPV2 activated by 2-aminoethoxydiphenyl borate, RR is tightly coordinated in the open selectivity filter, blocking ion flow and preventing channel inactivation. In TRPV5 activated by phosphatidylinositol 4,5-bisphosphate, RR blocks the selectivity filter and closes the lower gate through an interaction with polar residues in the pore vestibule. Together, our results provide a detailed understanding of TRPV subfamily pore block, the dynamic nature of the selectivity filter and allosteric communication between the selectivity filter and lower gate. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29051.map.gz emd_29051.map.gz | 85.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29051-v30.xml emd-29051-v30.xml emd-29051.xml emd-29051.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29051.png emd_29051.png | 136.5 KB | ||
| Filedesc metadata |  emd-29051.cif.gz emd-29051.cif.gz | 6.3 KB | ||
| Others |  emd_29051_half_map_1.map.gz emd_29051_half_map_1.map.gz emd_29051_half_map_2.map.gz emd_29051_half_map_2.map.gz | 66.8 MB 66.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29051 http://ftp.pdbj.org/pub/emdb/structures/EMD-29051 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29051 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29051 | HTTPS FTP |
-Related structure data
| Related structure data |  8ffqMC  8fflC  8ffmC  8ffnC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29051.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29051.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_29051_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
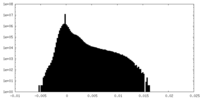
| Projections & Slices |
| ||||||||||||
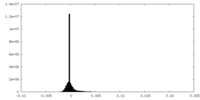
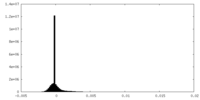
| Density Histograms |
-Half map: #1
| File | emd_29051_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Tetramer of wildtype rabbit TRPV5 into nanodiscs in complex with ...
| Entire | Name: Tetramer of wildtype rabbit TRPV5 into nanodiscs in complex with ruthenium red and PI(4,5)P2 |
|---|---|
| Components |
|
-Supramolecule #1: Tetramer of wildtype rabbit TRPV5 into nanodiscs in complex with ...
| Supramolecule | Name: Tetramer of wildtype rabbit TRPV5 into nanodiscs in complex with ruthenium red and PI(4,5)P2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Oryctolagus cuniculus (rabbit) Oryctolagus cuniculus (rabbit) |
| Molecular weight | Theoretical: 335.14 KDa |
-Macromolecule #1: Transient receptor potential cation channel subfamily V member 5
| Macromolecule | Name: Transient receptor potential cation channel subfamily V member 5 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Oryctolagus cuniculus (rabbit) Oryctolagus cuniculus (rabbit) |
| Molecular weight | Theoretical: 83.784586 KDa |
| Recombinant expression | Organism:   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) |
| Sequence | String: MGACPPKAKG PWAQLQKLLI SWPVGEQDWE QYRDRVNMLQ QERIRDSPLL QAAKENDLRL LKILLLNQSC DFQQRGAVGE TALHVAALY DNLEAATLLM EAAPELAKEP ALCEPFVGQT ALHIAVMNQN LNLVRALLAR GASVSARATG AAFRRSPHNL I YYGEHPLS ...String: MGACPPKAKG PWAQLQKLLI SWPVGEQDWE QYRDRVNMLQ QERIRDSPLL QAAKENDLRL LKILLLNQSC DFQQRGAVGE TALHVAALY DNLEAATLLM EAAPELAKEP ALCEPFVGQT ALHIAVMNQN LNLVRALLAR GASVSARATG AAFRRSPHNL I YYGEHPLS FAACVGSEEI VRLLIEHGAD IRAQDSLGNT VLHILILQPN KTFACQMYNL LLSYDEHSDH LQSLELVPNH QG LTPFKLA GVEGNTVMFQ HLMQKRKHVQ WTCGPLTSTL YDLTEIDSWG EELSFLELVV SSKKREARQI LEQTPVKELV SFK WKKYGR PYFCVLASLY ILYMICFTTC CIYRPLKLRD DNRTDPRDIT ILQQKLLQEA YVTHQDNIRL VGELVTVTGA VIIL LLEIP DIFRVGASRY FGQTILGGPF HVIIITYASL VLLTMVMRLT NMNGEVVPLS FALVLGWCSV MYFARGFQML GPFTI MIQK MIFGDLMRFC WLMAVVILGF ASAFHITFQT EDPNNLGEFS DYPTALFSTF ELFLTIIDGP ANYSVDLPFM YCITYA AFA IIATLLMLNL FIAMMGDTHW RVAQERDELW RAQVVATTVM LERKMPRFLW PRSGICGYEY GLGDRWFLRV ENHHDQN PL RVLRYVEAFK CSDKEDGQEQ LSEKRPSTVE SGMLSRASVA FQTPSLSRTT SQSSNSHRGW EILRRNTLGH LNLGLDLG E GDGEEVYHFT ETSQVAPA UniProtKB: Transient receptor potential cation channel subfamily V member 5 |
-Macromolecule #2: ERGOSTEROL
| Macromolecule | Name: ERGOSTEROL / type: ligand / ID: 2 / Number of copies: 8 / Formula: ERG |
|---|---|
| Molecular weight | Theoretical: 396.648 Da |
| Chemical component information |  ChemComp-ERG: |
-Macromolecule #3: ruthenium(6+) azanide pentaamino(oxido)ruthenium (1/4/2)
| Macromolecule | Name: ruthenium(6+) azanide pentaamino(oxido)ruthenium (1/4/2) type: ligand / ID: 3 / Number of copies: 1 / Formula: R2R |
|---|---|
| Molecular weight | Theoretical: 559.525 Da |
| Chemical component information | 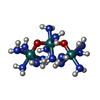 ChemComp-R2R: |
-Macromolecule #4: 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 4 / Number of copies: 4 / Formula: CPL |
|---|---|
| Molecular weight | Theoretical: 758.06 Da |
| Chemical component information |  ChemComp-CPL: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 105000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 12412 / Average electron dose: 43.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 6859887 |
|---|---|
| Startup model | Type of model: EMDB MAP EMDB ID: |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 3.3.2) |
| Final 3D classification | Number classes: 3 / Avg.num./class: 145000 / Software - Name: RELION (ver. 4.0) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 4.0) |
| Final reconstruction | Number classes used: 1 / Applied symmetry - Point group: C4 (4 fold cyclic ) / Resolution.type: BY AUTHOR / Resolution: 2.65 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 3.3.2) / Number images used: 368030 ) / Resolution.type: BY AUTHOR / Resolution: 2.65 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 3.3.2) / Number images used: 368030 |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8ffq: |
 Movie
Movie Controller
Controller








 Z
Z Y
Y X
X