+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 spike protein complexed with two nanobodies | |||||||||
 Map data Map data | Composite map (made from other two submitted maps) of nanobodies bound to SARS-CoV-2 spike protein. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  SARS-CoV-2 / spike / SARS-CoV-2 / spike /  nanobody / nanobody /  antibody / antibody /  VIRAL PROTEIN-IMMUNE SYSTEM complex VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationMaturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell ...Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / receptor-mediated endocytosis of virus by host cell / Attachment and Entry /  membrane fusion / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / membrane fusion / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell /  receptor ligand activity / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / receptor ligand activity / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / viral envelope / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane /  membrane / identical protein binding / membrane / identical protein binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Severe acute respiratory syndrome coronavirus 2 / Severe acute respiratory syndrome coronavirus 2 /   Lama glama (llama) Lama glama (llama) | |||||||||
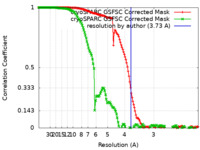
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.73 Å cryo EM / Resolution: 3.73 Å | |||||||||
 Authors Authors | Laughlin ZT / Patel A / Ortlund EA | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: SARS-CoV-2 spike protein bound with nanobodies Authors: Laughlin ZT / Patel A / Ortlund EA | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28686.map.gz emd_28686.map.gz | 255.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28686-v30.xml emd-28686-v30.xml emd-28686.xml emd-28686.xml | 28.2 KB 28.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28686_fsc.xml emd_28686_fsc.xml emd_28686_fsc_2.xml emd_28686_fsc_2.xml | 13.8 KB 13.8 KB | Display Display |  FSC data file FSC data file |
| Images |  emd_28686.png emd_28686.png | 143.5 KB | ||
| Filedesc metadata |  emd-28686.cif.gz emd-28686.cif.gz | 7.2 KB | ||
| Others |  emd_28686_additional_1.map.gz emd_28686_additional_1.map.gz emd_28686_additional_2.map.gz emd_28686_additional_2.map.gz emd_28686_additional_3.map.gz emd_28686_additional_3.map.gz emd_28686_additional_4.map.gz emd_28686_additional_4.map.gz emd_28686_additional_5.map.gz emd_28686_additional_5.map.gz emd_28686_additional_6.map.gz emd_28686_additional_6.map.gz | 258.8 MB 259.4 MB 254.6 MB 254.6 MB 254.7 MB 254.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28686 http://ftp.pdbj.org/pub/emdb/structures/EMD-28686 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28686 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28686 | HTTPS FTP |
-Related structure data
| Related structure data |  8eygMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28686.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28686.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite map (made from other two submitted maps) of nanobodies bound to SARS-CoV-2 spike protein. | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0691 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Local-refined map of nanobodies bound to SARS-CoV-2 spike protein.
| File | emd_28686_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local-refined map of nanobodies bound to SARS-CoV-2 spike protein. | ||||||||||||
| Projections & Slices |
| ||||||||||||
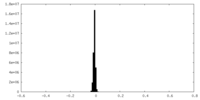
| Density Histograms |
-Additional map: Original, overall map of nanobodies bound to SARS-CoV-2...
| File | emd_28686_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Original, overall map of nanobodies bound to SARS-CoV-2 spike protein. | ||||||||||||
| Projections & Slices |
| ||||||||||||
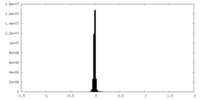
| Density Histograms |
-Additional map: Half-map of original, overall map of nanobodies bound...
| File | emd_28686_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map of original, overall map of nanobodies bound to SARS-CoV-2 spike protein. | ||||||||||||
| Projections & Slices |
| ||||||||||||
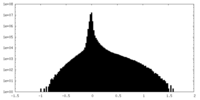
| Density Histograms |
-Additional map: Half-map of original, overall map of nanobodies bound...
| File | emd_28686_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map of original, overall map of nanobodies bound to SARS-CoV-2 spike protein. | ||||||||||||
| Projections & Slices |
| ||||||||||||
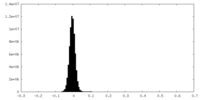
| Density Histograms |
-Additional map: Half-map of original, overall map of nanobodies bound...
| File | emd_28686_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map of original, overall map of nanobodies bound to SARS-CoV-2 spike protein. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Half-map of original, overall map of nanobodies bound...
| File | emd_28686_additional_6.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map of original, overall map of nanobodies bound to SARS-CoV-2 spike protein. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 spike protein trimer with two spike protein-targeting ...
| Entire | Name: SARS-CoV-2 spike protein trimer with two spike protein-targeting nanobodies attached on RBD domains. |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 spike protein trimer with two spike protein-targeting ...
| Supramolecule | Name: SARS-CoV-2 spike protein trimer with two spike protein-targeting nanobodies attached on RBD domains. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Severe acute respiratory syndrome coronavirus 2 Severe acute respiratory syndrome coronavirus 2 |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Severe acute respiratory syndrome coronavirus 2 Severe acute respiratory syndrome coronavirus 2 |
| Molecular weight | Theoretical: 125.882547 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QCVNLTTRTQ LPPAYTNSFT RGVYYPDKVF RSSVLHSTQD LFLPFFSNVT WFHAIHVSGT NGTKRFDNPV LPFNDGVYFA STEKSNIIR GWIFGTTLDS KTQSLLIVNN ATNVVIKVCE FQFCNDPFLG VYYHKNNKSW MESEFRVYSS ANNCTFEYVS Q PFLMDLEG ...String: QCVNLTTRTQ LPPAYTNSFT RGVYYPDKVF RSSVLHSTQD LFLPFFSNVT WFHAIHVSGT NGTKRFDNPV LPFNDGVYFA STEKSNIIR GWIFGTTLDS KTQSLLIVNN ATNVVIKVCE FQFCNDPFLG VYYHKNNKSW MESEFRVYSS ANNCTFEYVS Q PFLMDLEG KQGNFKNLRE FVFKNIDGYF KIYSKHTPIN LVRDLPQGFS ALEPLVDLPI GINITRFQTL LALHRSYLTP GD SSSGWTA GAAAYYVGYL QPRTFLLKYN ENGTITDAVD CALDPLSETK CTLKSFTVEK GIYQTSNFRV QPTESIVRFP NIT NLCPFG EVFNATRFAS VYAWNRKRIS NCVADYSVLY NSASFSTFKC YGVSPTKLND LCFTNVYADS FVIRGDEVRQ IAPG QTGKI ADYNYKLPDD FTGCVIAWNS NNLDSKVGGN YNYLYRLFRK SNLKPFERDI STEIYQAGST PCNGVEGFNC YFPLQ SYGF QPTNGVGYQP YRVVVLSFEL LHAPATVCGP KKSTNLVKNK CVNFNFNGLT GTGVLTESNK KFLPFQQFGR DIADTT DAV RDPQTLEILD ITPCSFGGVS VITPGTNTSN QVAVLYQDVN CTEVPVAIHA DQLTPTWRVY STGSNVFQTR AGCLIGA EH VNNSYECDIP IGAGICASYQ TQTNSPRRAR SVASQSIIAY TMSLGAENSV AYSNNSIAIP TNFTISVTTE ILPVSMTK T SVDCTMYICG DSTECSNLLL QYGSFCTQLN RALTGIAVEQ DKNTQEVFAQ VKQIYKTPPI KDFGGFNFSQ ILPDPSKPS KRSPIEDLLF NKVTLADAGF IKQYGDCLGD IAARDLICAQ KFNGLTVLPP LLTDEMIAQY TSALLAGTIT SGWTFGAGPA LQIPFPMQM AYRFNGIGVT QNVLYENQKL IANQFNSAIG KIQDSLSSTP SALGKLQDVV NQNAQALNTL VKQLSSNFGA I SSVLNDIL SRLDPPEAEV QIDRLITGRL QSLQTYVTQQ LIRAAEIRAS ANLAATKMSE CVLGQSKRVD FCGKGYHLMS FP QSAPHGV VFLHVTYVPA QEKNFTTAPA ICHDGKAHFP REGVFVSNGT HWFVTQRNFY EPQIITTDNT FVSGNCDVVI GIV NNTVYD PLQPELDSFK UniProtKB:  Spike glycoprotein Spike glycoprotein |
-Macromolecule #2: Nanobody
| Macromolecule | Name: Nanobody / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Lama glama (llama) Lama glama (llama) |
| Molecular weight | Theoretical: 12.360667 KDa |
| Recombinant expression | Organism:   Lama glama (llama) Lama glama (llama) |
| Sequence | String: EVQLVESGGG LVQPGGSLRL SCAASGGTFS SIGMGWFRQA PGKEREFVAA ISWDGGATAY ADSVKGRFTI SADNSKNTAY LQMNSLKPE DTAVYYCAKE DVGKPFDWGQ GTLVTVSSG |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 25 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 297 K / Instrument: FEI VITROBOT MARK IV / Details: Wait time 20 seconds and blot time 3 seconds. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 81000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 81000 |
| Specialist optics | Energy filter - Slit width: 30 eV |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 4186 / Average electron dose: 63.81 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 102 / Target criteria: Correlation coefficient |
|---|---|
| Output model |  PDB-8eyg: |
 Movie
Movie Controller
Controller












 Z
Z Y
Y X
X