[English] 日本語
 Yorodumi
Yorodumi- EMDB-28639: Cryo-EM structure of glutamate dehydrogenase frozen at various te... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of glutamate dehydrogenase frozen at various temperature | |||||||||
 Map data Map data | The EM map of Glutamate dehydrogenase (GDH) embedded in vitreous ice frozen at -183 celsius degree using standard procedure. The resolution of EM map is 2.7 angstrom | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationglutamate dehydrogenase [NAD(P)+] activity / glutamate catabolic process / tricarboxylic acid metabolic process / glutamate dehydrogenase [NAD(P)+] /  glutamate dehydrogenase (NADP+) activity / glutamate dehydrogenase (NAD+) activity / glutamine metabolic process / glutamate dehydrogenase (NADP+) activity / glutamate dehydrogenase (NAD+) activity / glutamine metabolic process /  mitochondrial inner membrane / GTP binding / mitochondrial inner membrane / GTP binding /  endoplasmic reticulum ...glutamate dehydrogenase [NAD(P)+] activity / glutamate catabolic process / tricarboxylic acid metabolic process / glutamate dehydrogenase [NAD(P)+] / endoplasmic reticulum ...glutamate dehydrogenase [NAD(P)+] activity / glutamate catabolic process / tricarboxylic acid metabolic process / glutamate dehydrogenase [NAD(P)+] /  glutamate dehydrogenase (NADP+) activity / glutamate dehydrogenase (NAD+) activity / glutamine metabolic process / glutamate dehydrogenase (NADP+) activity / glutamate dehydrogenase (NAD+) activity / glutamine metabolic process /  mitochondrial inner membrane / GTP binding / mitochondrial inner membrane / GTP binding /  endoplasmic reticulum / endoplasmic reticulum /  mitochondrion / mitochondrion /  ATP binding / identical protein binding ATP binding / identical protein bindingSimilarity search - Function | |||||||||
| Biological species |   Bos taurus (cattle) / Bos taurus (cattle) /   cattle (cattle) cattle (cattle) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.7 Å cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Shi H / Wu C / Zhang X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2023 Journal: Structure / Year: 2023Title: Addressing compressive deformation of proteins embedded in crystalline ice. Authors: Huigang Shi / Chunling Wu / Xinzheng Zhang /  Abstract: For cryoelectron microscopy (cryo-EM), high cooling rates have been required for preparation of protein samples to vitrify the surrounding water and avoid formation of damaging crystalline ice. ...For cryoelectron microscopy (cryo-EM), high cooling rates have been required for preparation of protein samples to vitrify the surrounding water and avoid formation of damaging crystalline ice. Whether and how crystalline ice affects single-particle cryo-EM is still unclear. Here, single-particle cryo-EM was used to analyze three-dimensional structures of various proteins and viruses embedded in crystalline ice formed at various cooling rates. Low cooling rates led to shrinkage deformation and density distortions on samples having loose structures. Higher cooling rates reduced deformations. Deformation-free proteins in crystalline ice were obtained by modifying the freezing conditions, and reconstructions from these samples revealed a marked improvement over vitreous ice. This procedure also increased the efficiency of cryo-EM structure determinations and was essential for high-resolution reconstructions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28639.map.gz emd_28639.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28639-v30.xml emd-28639-v30.xml emd-28639.xml emd-28639.xml | 45.2 KB 45.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28639.png emd_28639.png | 36.4 KB | ||
| Masks |  emd_28639_msk_1.map emd_28639_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_28639_additional_1.map.gz emd_28639_additional_1.map.gz emd_28639_additional_10.map.gz emd_28639_additional_10.map.gz emd_28639_additional_11.map.gz emd_28639_additional_11.map.gz emd_28639_additional_12.map.gz emd_28639_additional_12.map.gz emd_28639_additional_13.map.gz emd_28639_additional_13.map.gz emd_28639_additional_2.map.gz emd_28639_additional_2.map.gz emd_28639_additional_3.map.gz emd_28639_additional_3.map.gz emd_28639_additional_4.map.gz emd_28639_additional_4.map.gz emd_28639_additional_5.map.gz emd_28639_additional_5.map.gz emd_28639_additional_6.map.gz emd_28639_additional_6.map.gz emd_28639_additional_7.map.gz emd_28639_additional_7.map.gz emd_28639_additional_8.map.gz emd_28639_additional_8.map.gz emd_28639_additional_9.map.gz emd_28639_additional_9.map.gz emd_28639_half_map_1.map.gz emd_28639_half_map_1.map.gz emd_28639_half_map_2.map.gz emd_28639_half_map_2.map.gz | 59.9 MB 49.6 MB 49.6 MB 59.8 MB 49.5 MB 60 MB 49 MB 49 MB 59.9 MB 49.5 MB 49.6 MB 59 MB 59.8 MB 49 MB 49 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28639 http://ftp.pdbj.org/pub/emdb/structures/EMD-28639 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28639 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28639 | HTTPS FTP |
-Related structure data
| Related structure data |  8ew0MC  8bqnC  8ew2C  8f49C  8f7yC  8hhsC  8hi2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28639.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28639.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The EM map of Glutamate dehydrogenase (GDH) embedded in vitreous ice frozen at -183 celsius degree using standard procedure. The resolution of EM map is 2.7 angstrom | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||
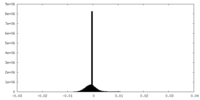
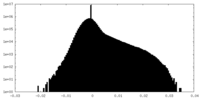
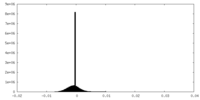
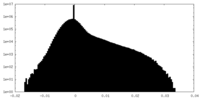
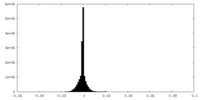
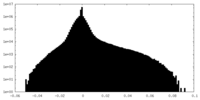
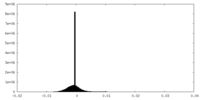
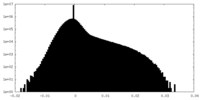
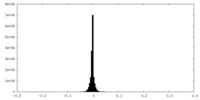
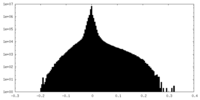
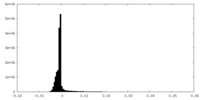
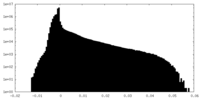
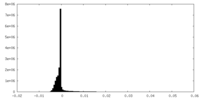
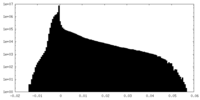
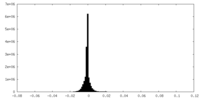
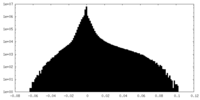
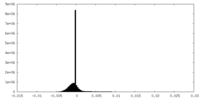
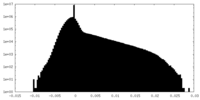
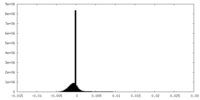
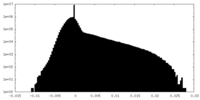
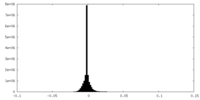
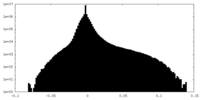
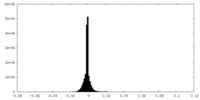
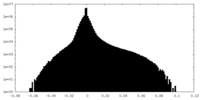
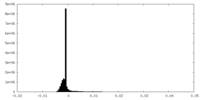
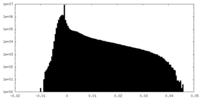
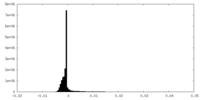
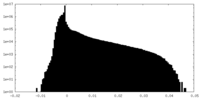
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
+Mask #1
+Additional map: The EM map of Glutamate dehydrogenase (GDH) embedded...
+Additional map: The half map of Glutamate dehydrogenase (GDH) embedded...
+Additional map: The half map of Glutamate dehydrogenase (GDH) embedded...
+Additional map: The EM map of Glutamate dehydrogenase (GDH) embedded...
+Additional map: The half map of Glutamate dehydrogenase (GDH) embedded...
+Additional map: The EM map of Glutamate dehydrogenase (GDH) embedded...
+Additional map: The half map of Glutamate dehydrogenase (GDH) embedded...
+Additional map: The half map of Glutamate dehydrogenase (GDH) embedded...
+Additional map: The EM map of Glutamate dehydrogenase (GDH) embedded...
+Additional map: The half map of Glutamate dehydrogenase (GDH) embedded...
+Additional map: The half map of Glutamate dehydrogenase (GDH) embedded...
+Additional map: The EM map of Glutamate dehydrogenase (GDH) embedded...
+Additional map: The EM map of Glutamate dehydrogenase (GDH) embedded...
+Half map: The half map of Glutamate dehydrogenase (GDH) embedded...
+Half map: The EM map of Glutamate dehydrogenase (GDH) embedded...
- Sample components
Sample components
-Entire : Glutamate Dehydrogenase
| Entire | Name: Glutamate Dehydrogenase |
|---|---|
| Components |
|
-Supramolecule #1: Glutamate Dehydrogenase
| Supramolecule | Name: Glutamate Dehydrogenase / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Bos taurus (cattle) Bos taurus (cattle) |
-Macromolecule #1: Glutamate dehydrogenase 1, mitochondrial
| Macromolecule | Name: Glutamate dehydrogenase 1, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: glutamate dehydrogenase [NAD(P)+] |
|---|---|
| Source (natural) | Organism:   cattle (cattle) cattle (cattle) |
| Molecular weight | Theoretical: 61.60891 KDa |
| Sequence | String: MYRYLGEALL LSRAGPAALG SASADSAALL GWARGQPAAA PQPGLVPPAR RHYSEAAADR EDDPNFFKMV EGFFDRGASI VEDKLVEDL KTRETEEQKR NRVRSILRII KPCNHVLSLS FPIRRDDGSW EVIEGYRAQH SQHRTPCKGG IRYSTDVSVD E VKALASLM ...String: MYRYLGEALL LSRAGPAALG SASADSAALL GWARGQPAAA PQPGLVPPAR RHYSEAAADR EDDPNFFKMV EGFFDRGASI VEDKLVEDL KTRETEEQKR NRVRSILRII KPCNHVLSLS FPIRRDDGSW EVIEGYRAQH SQHRTPCKGG IRYSTDVSVD E VKALASLM TYKCAVVDVP FGGAKAGVKI NPKNYTDNEL EKITRRFTME LAKKGFIGPG VDVPAPDMST GEREMSWIAD TY ASTIGHY DINAHACVTG KPISQGGIHG RISATGRGVF HGIENFINEA SYMSILGMTP GFGDKTFVVQ GFGNVGLHSM RYL HRFGAK CITVGESDGS IWNPDGIDPK ELEDFKLQHG TILGFPKAKI YEGSILEVDC DILIPAASEK QLTKSNAPRV KAKI IAEGA NGPTTPEADK IFLERNIMVI PDLYLNAGGV TVSYFEWLKN LNHVSYGRLT FKYERDSNYH LLMSVQESLE RKFGK HGGT IPIVPTAEFQ DRISGASEKD IVHSGLAYTM ERSARQIMRT AMKYNLGLDL RTAAYVNAIE KVFRVYNEAG VTFT |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 60.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION |
|---|---|
| Final 3D classification | Software - Name: RELION |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.7 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 122399 |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-8ew0: |
 Movie
Movie Controller
Controller









 Z
Z Y
Y X
X