[English] 日本語
 Yorodumi
Yorodumi- EMDB-28256: 30S-focused map for: "Atomistic simulations of the E. coli riboso... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
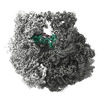
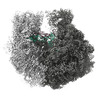
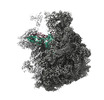
| Title | 30S-focused map for: "Atomistic simulations of the E. coli ribosome provide selection criteria for translationally active substrates" | |||||||||
 Map data Map data | Post-processed, masked 30S-focused map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  ribosome / ribosome /  tRNA / tRNA /  e. coli e. coli | |||||||||
| Biological species |   Escherichia coli (E. coli) Escherichia coli (E. coli) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.3 Å cryo EM / Resolution: 2.3 Å | |||||||||
 Authors Authors | Watson ZL / Cate JHD | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Chem / Year: 2023 Journal: Nat Chem / Year: 2023Title: Atomistic simulations of the Escherichia coli ribosome provide selection criteria for translationally active substrates. Authors: Zoe L Watson / Isaac J Knudson / Fred R Ward / Scott J Miller / Jamie H D Cate / Alanna Schepartz / Ara M Abramyan /  Abstract: As genetic code expansion advances beyond L-α-amino acids to backbone modifications and new polymerization chemistries, delineating what substrates the ribosome can accommodate remains a challenge. ...As genetic code expansion advances beyond L-α-amino acids to backbone modifications and new polymerization chemistries, delineating what substrates the ribosome can accommodate remains a challenge. The Escherichia coli ribosome tolerates non-L-α-amino acids in vitro, but few structural insights that explain how are available, and the boundary conditions for efficient bond formation are so far unknown. Here we determine a high-resolution cryogenic electron microscopy structure of the E. coli ribosome containing α-amino acid monomers and use metadynamics simulations to define energy surface minima and understand incorporation efficiencies. Reactive monomers across diverse structural classes favour a conformational space where the aminoacyl-tRNA nucleophile is <4 Å from the peptidyl-tRNA carbonyl with a Bürgi-Dunitz angle of 76-115°. Monomers with free energy minima that fall outside this conformational space do not react efficiently. This insight should accelerate the in vivo and in vitro ribosomal synthesis of sequence-defined, non-peptide heterooligomers. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28256.map.gz emd_28256.map.gz | 30.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28256-v30.xml emd-28256-v30.xml emd-28256.xml emd-28256.xml | 22.3 KB 22.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28256_fsc.xml emd_28256_fsc.xml | 16.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_28256.png emd_28256.png | 104.4 KB | ||
| Masks |  emd_28256_msk_1.map emd_28256_msk_1.map | 361.7 MB |  Mask map Mask map | |
| Others |  emd_28256_additional_1.map.gz emd_28256_additional_1.map.gz emd_28256_half_map_1.map.gz emd_28256_half_map_1.map.gz emd_28256_half_map_2.map.gz emd_28256_half_map_2.map.gz | 288.4 MB 289.6 MB 289.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28256 http://ftp.pdbj.org/pub/emdb/structures/EMD-28256 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28256 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28256 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28256.map.gz / Format: CCP4 / Size: 361.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28256.map.gz / Format: CCP4 / Size: 361.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post-processed, masked 30S-focused map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8296 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_28256_msk_1.map emd_28256_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Map from 30S-focused 3D auto-refine, without post-processing
| File | emd_28256_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map from 30S-focused 3D auto-refine, without post-processing | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unfiltered half-map 1
| File | emd_28256_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unfiltered half-map 2
| File | emd_28256_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : 30S-focused map
| Entire | Name: 30S-focused map |
|---|---|
| Components |
|
-Supramolecule #1: 30S-focused map
| Supramolecule | Name: 30S-focused map / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#54 Details: Overall complex includes 70S ribosome with mRNA, A- and P-site Met-NH-tRNAs |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X



































