[English] 日本語
 Yorodumi
Yorodumi- EMDB-27981: SsoMCM hexamer bound to Mg/ADP-BeFx and DNA. Class 2. Merged part... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SsoMCM hexamer bound to Mg/ADP-BeFx and DNA. Class 2. Merged particles from datasets with 3 different DNA entities | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationMCM complex / DNA duplex unwinding /  helicase activity / helicase activity /  single-stranded DNA binding / single-stranded DNA binding /  DNA helicase / DNA helicase /  DNA replication / DNA replication /  ATP hydrolysis activity / ATP hydrolysis activity /  ATP binding / identical protein binding ATP binding / identical protein bindingSimilarity search - Function | |||||||||
| Biological species |   Saccharolobus solfataricus P2 (archaea) / synthetic construct (others) Saccharolobus solfataricus P2 (archaea) / synthetic construct (others) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.59 Å cryo EM / Resolution: 2.59 Å | |||||||||
 Authors Authors | Meagher M / Myasnikov A / Enemark EJ | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Int J Mol Sci / Year: 2022 Journal: Int J Mol Sci / Year: 2022Title: Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation. Authors: Martin Meagher / Alexander Myasnikov / Eric J Enemark /  Abstract: A six-subunit ATPase ring forms the central hub of the replication forks in all domains of life. This ring performs a helicase function to separate the two complementary DNA strands to be replicated ...A six-subunit ATPase ring forms the central hub of the replication forks in all domains of life. This ring performs a helicase function to separate the two complementary DNA strands to be replicated and drives the replication machinery along the DNA. Disruption of this helicase/ATPase ring is associated with genetic instability and diseases such as cancer. The helicase/ATPase rings of eukaryotes and archaea consist of six minichromosome maintenance (MCM) proteins. Prior structural studies have shown that MCM rings bind one encircled strand of DNA in a spiral staircase, suggesting that the ring pulls this strand of DNA through its central pore in a hand-over-hand mechanism where the subunit at the bottom of the staircase dissociates from DNA and re-binds DNA one step above the staircase. With high-resolution cryo-EM, we show that the MCM ring of the archaeal organism binds an encircled DNA strand in two different modes with different numbers of subunits engaged to DNA, illustrating a plausible mechanism for the alternating steps of DNA dissociation and re-association that occur during DNA translocation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27981.map.gz emd_27981.map.gz | 31.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27981-v30.xml emd-27981-v30.xml emd-27981.xml emd-27981.xml | 22.7 KB 22.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27981_fsc.xml emd_27981_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_27981.png emd_27981.png | 118.6 KB | ||
| Masks |  emd_27981_msk_1.map emd_27981_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_27981_additional_1.map.gz emd_27981_additional_1.map.gz emd_27981_half_map_1.map.gz emd_27981_half_map_1.map.gz emd_27981_half_map_2.map.gz emd_27981_half_map_2.map.gz | 59.7 MB 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27981 http://ftp.pdbj.org/pub/emdb/structures/EMD-27981 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27981 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27981 | HTTPS FTP |
-Related structure data
| Related structure data |  8eamMC  8eafC  8eagC  8eahC  8eaiC  8eajC  8eakC  8ealC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27981.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27981.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27981_msk_1.map emd_27981_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
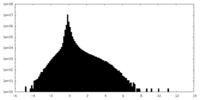
| Density Histograms |
-Additional map: sharpened map
| File | emd_27981_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
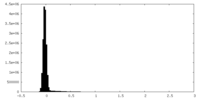
| Density Histograms |
-Half map: half-map A
| File | emd_27981_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
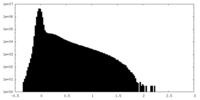
| Density Histograms |
-Half map: half-map B
| File | emd_27981_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
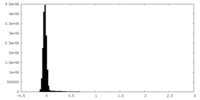
| Density Histograms |
- Sample components
Sample components
-Entire : SsoMCM hexamer bound to Mg/ADP-BeFx and DNA. Class 2. Merged part...
| Entire | Name: SsoMCM hexamer bound to Mg/ADP-BeFx and DNA. Class 2. Merged particles from datasets with 3 different DNA entities. |
|---|---|
| Components |
|
-Supramolecule #1: SsoMCM hexamer bound to Mg/ADP-BeFx and DNA. Class 2. Merged part...
| Supramolecule | Name: SsoMCM hexamer bound to Mg/ADP-BeFx and DNA. Class 2. Merged particles from datasets with 3 different DNA entities. type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Saccharolobus solfataricus P2 (archaea) Saccharolobus solfataricus P2 (archaea) |
-Macromolecule #1: Minichromosome maintenance protein MCM
| Macromolecule | Name: Minichromosome maintenance protein MCM / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number:  DNA helicase DNA helicase |
|---|---|
| Source (natural) | Organism:   Saccharolobus solfataricus P2 (archaea) / Strain: ATCC 35092 / DSM 1617 / JCM 11322 / P2 Saccharolobus solfataricus P2 (archaea) / Strain: ATCC 35092 / DSM 1617 / JCM 11322 / P2 |
| Molecular weight | Theoretical: 68.641961 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: SLEIPSKQID YRDVFIEFLT TFKGNNNQNK YIERINELVA YRKKSLIIEF SDVLSFNENL AYEIINNTKI ILPILEGALY DHILQLDPT YQRDIEKVHV RIVGIPRVIE LRKIRSTDIG KLITIDGILV KVTPVKERIY KATYKHIHPD CMQEFEWPED E EMPEVLEM ...String: SLEIPSKQID YRDVFIEFLT TFKGNNNQNK YIERINELVA YRKKSLIIEF SDVLSFNENL AYEIINNTKI ILPILEGALY DHILQLDPT YQRDIEKVHV RIVGIPRVIE LRKIRSTDIG KLITIDGILV KVTPVKERIY KATYKHIHPD CMQEFEWPED E EMPEVLEM PTICPKCGKP GQFRLIPEKT KLIDWQKAVI QERPEEVPSG QLPRQLEIIL EDDLVDSARP GDRVKVTGIL DI KQDSPVK RGSRAVFDIY MKVSSIEVSG GSGGSSEEDE KKIKDLAKDP WIRDRIISSI APSIYGHWEL KEALALALFG GVP KVLEDT RIRGDIHILI IGDPGTAKSQ MLQFISRVAP RAVYTTGKGS TAAGLTAAVV REKGTGEYYL EAGALVLADG GIAV IDEID KMRDEDRVAI HEAMEQQTVS IAKAGIVAKL NARAAVIAAG NPKFGRYISE RPVSDNINLP PTILSRFDLI FILKD QPGE QDRELANYIL DVHSGKSTKN IIDIDTLRKY IAYARKYVTP KITSEAKNLI TDFFVEMRKK SSETPDSPIL ITPRQL EAL IRISEAYAKM ALKAEVTRED AERAINIMRL FLESVGVDME SGKIDID |
-Macromolecule #2: DNA
| Macromolecule | Name: DNA / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 3.605356 KDa |
| Sequence | String: (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT) |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 6 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: [[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-...
| Macromolecule | Name: [[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-tris(fluoranyl)beryllium type: ligand / ID: 5 / Number of copies: 4 / Formula: 08T |
|---|---|
| Molecular weight | Theoretical: 492.201 Da |
| Chemical component information |  ChemComp-08T: |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 12 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 78.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: B |
|---|---|
| Details | Bond distance and angle restraints for a tetrahedral Zn2+ coordination were applied. Bond distance and angle restraints for a octahedral Mg2+ coordination were applied. |
| Refinement | Space: REAL / Protocol: OTHER |
| Output model |  PDB-8eam: |
 Movie
Movie Controller
Controller











 Z
Z Y
Y X
X