[English] 日本語
 Yorodumi
Yorodumi- EMDB-27822: Structure of the human UBR5 HECT-type E3 ubiquitin ligase in a C2... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 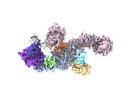 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the human UBR5 HECT-type E3 ubiquitin ligase in a C2 symmetric dimeric form | |||||||||
 Map data Map data | Final composite map | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationheterochromatin boundary formation / HECT-type E3 ubiquitin transferase / DNA repair-dependent chromatin remodeling / ubiquitin-ubiquitin ligase activity / progesterone receptor signaling pathway / protein K48-linked ubiquitination /  ubiquitin binding / protein polyubiquitination / positive regulation of protein import into nucleus / positive regulation of canonical Wnt signaling pathway ...heterochromatin boundary formation / HECT-type E3 ubiquitin transferase / DNA repair-dependent chromatin remodeling / ubiquitin-ubiquitin ligase activity / progesterone receptor signaling pathway / protein K48-linked ubiquitination / ubiquitin binding / protein polyubiquitination / positive regulation of protein import into nucleus / positive regulation of canonical Wnt signaling pathway ...heterochromatin boundary formation / HECT-type E3 ubiquitin transferase / DNA repair-dependent chromatin remodeling / ubiquitin-ubiquitin ligase activity / progesterone receptor signaling pathway / protein K48-linked ubiquitination /  ubiquitin binding / protein polyubiquitination / positive regulation of protein import into nucleus / positive regulation of canonical Wnt signaling pathway / ubiquitin binding / protein polyubiquitination / positive regulation of protein import into nucleus / positive regulation of canonical Wnt signaling pathway /  ubiquitin protein ligase activity / ubiquitin protein ligase activity /  DNA repair / DNA damage response / positive regulation of gene expression / perinuclear region of cytoplasm / protein-containing complex / DNA repair / DNA damage response / positive regulation of gene expression / perinuclear region of cytoplasm / protein-containing complex /  RNA binding / zinc ion binding / RNA binding / zinc ion binding /  nucleoplasm / nucleoplasm /  membrane / membrane /  nucleus / nucleus /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.66 Å cryo EM / Resolution: 2.66 Å | |||||||||
 Authors Authors | Wang F / He Q / Lin G / Li H | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2023 Journal: Structure / Year: 2023Title: Structure of the human UBR5 E3 ubiquitin ligase. Authors: Feng Wang / Qing He / Wenhu Zhan / Ziqi Yu / Efrat Finkin-Groner / Xiaojing Ma / Gang Lin / Huilin Li /  Abstract: The human UBR5 is a single polypeptide chain homology to E6AP C terminus (HECT)-type E3 ubiquitin ligase essential for embryonic development in mammals. Dysregulated UBR5 functions like an ...The human UBR5 is a single polypeptide chain homology to E6AP C terminus (HECT)-type E3 ubiquitin ligase essential for embryonic development in mammals. Dysregulated UBR5 functions like an oncoprotein to promote cancer growth and metastasis. Here, we report that UBR5 assembles into a dimer and a tetramer. Our cryoelectron microscopy (cryo-EM) structures reveal that two crescent-shaped UBR5 monomers assemble head to tail to form the dimer, and two dimers bind face to face to form the cage-like tetramer with all four catalytic HECT domains facing the central cavity. Importantly, the N-terminal region of one subunit and the HECT of the other form an "intermolecular jaw" in the dimer. We show the jaw-lining residues are important for function, suggesting that the intermolecular jaw functions to recruit ubiquitin-loaded E2 to UBR5. Further work is needed to understand how oligomerization regulates UBR5 ligase activity. This work provides a framework for structure-based anticancer drug development and contributes to a growing appreciation of E3 ligase diversity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27822.map.gz emd_27822.map.gz | 217.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27822-v30.xml emd-27822-v30.xml emd-27822.xml emd-27822.xml | 23.9 KB 23.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27822.png emd_27822.png | 65.7 KB | ||
| Others |  emd_27822_additional_1.map.gz emd_27822_additional_1.map.gz emd_27822_additional_2.map.gz emd_27822_additional_2.map.gz emd_27822_additional_3.map.gz emd_27822_additional_3.map.gz emd_27822_additional_4.map.gz emd_27822_additional_4.map.gz | 216.1 MB 216.2 MB 216.7 MB 118.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27822 http://ftp.pdbj.org/pub/emdb/structures/EMD-27822 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27822 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27822 | HTTPS FTP |
-Related structure data
| Related structure data |  8e0qMC  8d4xC  8ewiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27822.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27822.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final composite map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.828 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Focused on refinment map of UBR5-HECT
| File | emd_27822_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused on refinment map of UBR5-HECT | ||||||||||||
| Projections & Slices |
| ||||||||||||
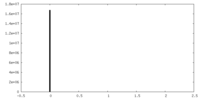
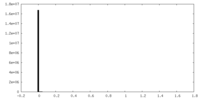
| Density Histograms |
-Additional map: Focused on refinment map of UBR5-NTR
| File | emd_27822_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused on refinment map of UBR5-NTR | ||||||||||||
| Projections & Slices |
| ||||||||||||
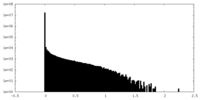
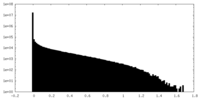
| Density Histograms |
-Additional map: sharpen map of UBR5 in C2 symmetry
| File | emd_27822_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpen map of UBR5 in C2 symmetry | ||||||||||||
| Projections & Slices |
| ||||||||||||
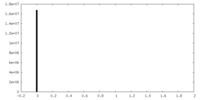
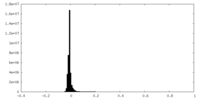
| Density Histograms |
-Additional map: Raw EM map of UBR5 in C2 symmetry
| File | emd_27822_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Raw EM map of UBR5 in C2 symmetry | ||||||||||||
| Projections & Slices |
| ||||||||||||
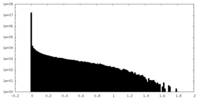
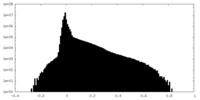
| Density Histograms |
- Sample components
Sample components
-Entire : Homodimer of human E3 ligase UBR5
| Entire | Name: Homodimer of human E3 ligase UBR5 |
|---|---|
| Components |
|
-Supramolecule #1: Homodimer of human E3 ligase UBR5
| Supramolecule | Name: Homodimer of human E3 ligase UBR5 / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1 / Details: UBR5 expressed in insect cell |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 610 KDa |
-Macromolecule #1: E3 ubiquitin-protein ligase UBR5
| Macromolecule | Name: E3 ubiquitin-protein ligase UBR5 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: HECT-type E3 ubiquitin transferase |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 310.719719 KDa |
| Recombinant expression | Organism: Insect cell expression vector pTIE1 (others) |
| Sequence | String: DYKDDDDKMT SIHFVVHPLP GTEDQLNDRL REVSEKLNKY NLNSHPPLNV LEQATIKQCV VGPNHAAFLL EDGRVCRIGF SVQPDRLEL GKPDNNDGSK LNSNSGAGRT SRPGRTSDSP WFLSGSETLG RLAGNTLGSR WSSGVGGSGG GSSGRSSAGA R DSRRQTRV ...String: DYKDDDDKMT SIHFVVHPLP GTEDQLNDRL REVSEKLNKY NLNSHPPLNV LEQATIKQCV VGPNHAAFLL EDGRVCRIGF SVQPDRLEL GKPDNNDGSK LNSNSGAGRT SRPGRTSDSP WFLSGSETLG RLAGNTLGSR WSSGVGGSGG GSSGRSSAGA R DSRRQTRV IRTGRDRGSG LLGSQPQPVI PASVIPEELI SQAQVVLQGK SRSVIIRELQ RTNLDVNLAV NNLLSRDDED GD DGDDTAS ESYLPGEDLM SLLDADIHSA HPSVIIDADA MFSEDISYFG YPSFRRSSLS RLGSSRVLLL PLERDSELLR ERE SVLRLR ERRWLDGASF DNERGSTSKE GEPNLDKKNT PVQSPVSLGE DLQWWPDKDG TKFICIGALY SELLAVSSKG ELYQ WKWSE SEPYRNAQNP SLHHPRATFL GLTNEKIVLL SANSIRATVA TENNKVATWV DETLSSVASK LEHTAQTYSE LQGER IVSL HCCALYTCAQ LENSLYWWGV VPFSQRKKML EKARAKNKKP KSSAGISSMP NITVGTQVCL RNNPLYHAGA VAFSIS AGI PKVGVLMESV WNMNDSCRFQ LRSPESLKNM EKASKTTEAK PESKQEPVKT EMGPPPSPAS TCSDASSIAS SASMPYK RR RSTPAPKEEE KVNEEQWSLR EVVFVEDVKN VPVGKVLKVD GAYVAVKFPG TSSNTNCQNS SGPDADPSSL LQDCRLLR I DELQVVKTGG TPKVPDCFQR TPKKLCIPEK TEILAVNVDS KGVHAVLKTG NWVRYCIFDL ATGKAEQENN FPTSSIAFL GQNERNVAIF TAGQESPIIL RDGNGTIYPM AKDCMGGIRD PDWLDLPPIS SLGMGVHSLI NLPANSTIKK KAAVIIMAVE KQTLMQHIL RCDYEACRQY LMNLEQAVVL EQNLQMLQTF ISHRCDGNRN ILHACVSVCF PTSNKETKEE EEAERSERNT F AERLSAVE AIANAISVVS SNGPGNRAGS SSSRSLRLRE MMRRSLRAAG LGRHEAGASS SDHQDPVSPP IAPPSWVPDP PA MDPDGDI DFILAPAVGS LTTAATGTGQ GPSTSTIPGP STEPSVVESK DRKANAHFIL KLLCDSVVLQ PYLRELLSAK DAR GMTPFM SAVSGRAYPA AITILETAQK IAKAEISSSE KEEDVFMGMV CPSGTNPDDS PLYVLCCNDT CSFTWTGAEH INQD IFECR TCGLLESLCC CTECARVCHK GHDCKLKRTS PTAYCDCWEK CKCKTLIAGQ KSARLDLLYR LLTATNLVTL PNSRG EHLL LFLVQTVARQ TVEHCQYRPP RIREDRNRKT ASPEDSDMPD HDLEPPRFAQ LALERVLQDW NALKSMIMFG SQENKD PLS ASSRIGHLLP EEQVYLNQQS GTIRLDCFTH CLIVKCTADI LLLDTLLGTL VKELQNKYTP GRREEAIAVT MRFLRSV AR VFVILSVEMA SSKKKNNFIP QPIGKCKRVF QALLPYAVEE LCNVAESLIV PVRMGIARPT APFTLASTSI DAMQGSEE L FSVEPLPPRP SSDQSSSSSQ SQSSYIIRNP QQRRISQSQP VRGRDEEQDD IVSADVEEVE VVEGVAGEED HHDEQEEHG EENAEAEGQH DEHDEDGSDM ELDLLAAAET ESDSESNHSN QDNASGRRSV VTAATAGSEA GASSVPAFFS EDDSQSNDSS DSDSSSSQS DDIEQETFML DEPLERTTNS SHANGAAQAP RSMQWAVRNT QHQRAASTAP SSTSTPAASS AGLIYIDPSN L RRSGTIST SAAAAAAALE ASNASSYLTS ASSLARAYSI VIRQISDLMG LIPKYNHLVY SQIPAAVKLT YQDAVNLQNY VE EKLIPTW NWMVSIMDST EAQLRYGSAL ASAGDPGHPN HPLHASQNSA RRERMTAREE ASLRTLEGRR RATLLSARQG MMS ARGDFL NYALSLMRSH NDEHSDVLPV LDVCSLKHVA YVFQALIYWI KAMNQQTTLD TPQLERKRTR ELLELGIDNE DSEH ENDDD TNQSATLNDK DDDSLPAETG QNHPFFRRSD SMTFLGCIPP NPFEVPLAEA IPLADQPHLL QPNARKEDLF GRPSQ GLYS SSASSGKCLM EVTVDRNCLE VLPTKMSYAA NLKNVMNMQN RQKKEGEEQP VLPEETESSK PGPSAHDLAA QLKSSL LAE IGLTESEGPP LTSFRPQCSF MGMVISHDML LGRWRLSLEL FGRVFMEDVG AEPGSILTEL GGFEVKESKF RREMEKL RN QQSRDLSLEV DRDRDLLIQQ TMRQLNNHFG RRCATTPMAV HRVKVTFKDE PGEGSGVARS FYTAIAQAFL SNEKLPNL E CIQNANKGTH TSLMQRLRNR GERDRERERE REMRRSSGLR AGSRRDRDRD FRRQLSIDTR PFRPASEGNP SDDPEPLPA HRQALGERLY PRVQAMQPAF ASKITGMLLE LSPAQLLLLL ASEDSLRARV DEAMELIIAH GRENGADSIL DLGLVDSSEK VQQENRKRH GSSRSVVDMD LDDTDDGDDN APLFYQPGKR GFYTPRPGKN TEARLNCFRN IGRILGLCLL QNELCPITLN R HVIKVLLG RKVNWHDFAF FDPVMYESLR QLILASQSSD ADAVFSAMDL AFAIDLCKEE GGGQVELIPN GVNIPVTPQN VY EYVRKYA EHRMLVVAEQ PLHAMRKGLL DVLPKNSLED LTAEDFRLLV NGCGEVNVQM LISFTSFNDE SGENAEKLLQ FKR WFWSIV EKMSMTERQD LVYFWTSSPS LPASEEGFQP MPSITIRPPD DQHLPTANTC ISRLYVPLYS SKQILKQKLL LAIK TKNFG FV |
-Macromolecule #2: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 2 / Number of copies: 6 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/1 / Support film - Material: GOLD / Support film - topology: CONTINUOUS | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 279 K / Instrument: FEI VITROBOT MARK IV / Details: blot 2S, blot forth 2. | ||||||||||||
| Details | freshly purified UBR5 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Temperature | Min: 193.0 K / Max: 193.0 K |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 65.0 e/Å2 Details: Images were collected in movie-mode at 75 frames per second |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
|---|---|
| Final 3D classification | Number classes: 3 / Avg.num./class: 80 |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.66 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 3.2.0) / Number images used: 844403 |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 119 / Target criteria: Correlation coefficient |
|---|---|
| Output model |  PDB-8e0q: |
 Movie
Movie Controller
Controller


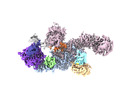
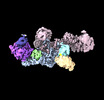
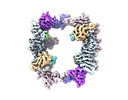

 Z
Z Y
Y X
X