+ Open data
Open data
- Basic information
Basic information
| Entry | 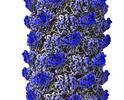 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
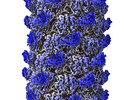
| Title | Tail of P1 bacteriophage | |||||||||
 Map data Map data | Tail of P1 bacteriophage | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | phage tail / helical /  cryo-TEM / cryo-TEM /  CONTRACTILE PROTEIN CONTRACTILE PROTEIN | |||||||||
| Biological species |   Escherichia phage P1 (virus) Escherichia phage P1 (virus) | |||||||||
| Method | helical reconstruction /  cryo EM / Resolution: 3.53 Å cryo EM / Resolution: 3.53 Å | |||||||||
 Authors Authors | Sen A / Bhatt V / Kim K / Bouchet-Marquis C | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Tail of P1 bacteriophage Authors: Sen A / Bhatt V / Kim K / Bouchet-Marquis C | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27714.map.gz emd_27714.map.gz | 483.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27714-v30.xml emd-27714-v30.xml emd-27714.xml emd-27714.xml | 13 KB 13 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27714.png emd_27714.png | 209.8 KB | ||
| Filedesc metadata |  emd-27714.cif.gz emd-27714.cif.gz | 4.7 KB | ||
| Others |  emd_27714_half_map_1.map.gz emd_27714_half_map_1.map.gz emd_27714_half_map_2.map.gz emd_27714_half_map_2.map.gz | 77.4 MB 77.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27714 http://ftp.pdbj.org/pub/emdb/structures/EMD-27714 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27714 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27714 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27714.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27714.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Tail of P1 bacteriophage | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.69445 Å | ||||||||||||||||||||
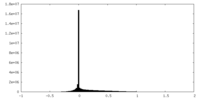
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map of Tail of P1 bacteriophage
| File | emd_27714_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map of Tail of P1 bacteriophage | ||||||||||||
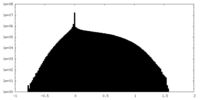
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map of Tail of P1 bacteriophage
| File | emd_27714_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map of Tail of P1 bacteriophage | ||||||||||||
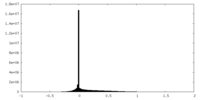
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Escherichia phage P1
| Entire | Name:   Escherichia phage P1 (virus) Escherichia phage P1 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Escherichia phage P1
| Supramolecule | Name: Escherichia phage P1 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2886926 / Sci species name: Escherichia phage P1 / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Phage tail protein
| Macromolecule | Name: Phage tail protein / type: other / ID: 1 / Classification: other |
|---|---|
| Source (natural) | Organism:   Escherichia phage P1 (virus) Escherichia phage P1 (virus) |
| Sequence | String: MQRSWFNHRL TSAKQKSLLY KSLADLVQSM MDTFVDPWLE RITN RKSIF SMSKEDLETR TNELGQFFTI RTSNSSSVPM LLQQRLDEIH FKGTERPINQ TIY REFNGI SVLWDPIYAP VDLERHPYGT VLIPESTLET TGGTFGEMFL TSRGMISIPI ND LARTMGI ...String: MQRSWFNHRL TSAKQKSLLY KSLADLVQSM MDTFVDPWLE RITN RKSIF SMSKEDLETR TNELGQFFTI RTSNSSSVPM LLQQRLDEIH FKGTERPINQ TIY REFNGI SVLWDPIYAP VDLERHPYGT VLIPESTLET TGGTFGEMFL TSRGMISIPI ND LARTMGI TGTIDQSAIT EEILRKFNQF VKPLLPLHIV FDGLTLYLSV VVNEHADMIT L NEISDTEK AYCWFETSDT TSLTGVTSIS APITATPGGT IVKATPTFDR TRADDLLLDS DA |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 73000 Bright-field microscopy / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 73000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average exposure time: 9.4 sec. / Average electron dose: 30.0 e/Å2 |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final angle assignment | Type: NOT APPLICABLE |
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 39.15 Å Applied symmetry - Helical parameters - Δ&Phi: 19.32 ° Applied symmetry - Helical parameters - Axial symmetry: C6 (6 fold cyclic  ) )Resolution.type: BY AUTHOR / Resolution: 3.53 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 119472 |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X