[English] 日本語
 Yorodumi
Yorodumi- EMDB-27431: Cryo-EM structure of Saccharomyces cerevisiae Succinyl-CoA:acetat... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Saccharomyces cerevisiae Succinyl-CoA:acetate CoA-transferase (Ach1p) | |||||||||
 Map data Map data | map sharp | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ach1p /  Succinyl-CoA / Succinyl-CoA /  acetate CoA-transferase / acetate CoA-transferase /  TRANSFERASE TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology information acetyl-CoA hydrolase / propionate metabolic process, methylcitrate cycle / acetyl-CoA hydrolase / propionate metabolic process, methylcitrate cycle /  acetyl-CoA hydrolase activity / acetyl-CoA hydrolase activity /  acetate CoA-transferase activity / acetate metabolic process / acetate CoA-transferase activity / acetate metabolic process /  mitochondrion / mitochondrion /  cytosol cytosolSimilarity search - Function | |||||||||
| Biological species |   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) | |||||||||
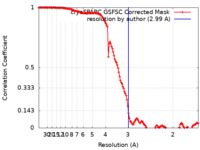
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.99 Å cryo EM / Resolution: 2.99 Å | |||||||||
 Authors Authors | Godoy AS / Song Y / Cheruvara H / Quigley A / Oliva G | |||||||||
| Funding support |  Brazil, 1 items Brazil, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of Saccharomyces cerevisiae cytochrome c oxidase (Complex IV) extracted in lipid nanodiscs Authors: Godoy AS / Song Y / Cheruvara H / Quigley A / Oliva G | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27431.map.gz emd_27431.map.gz | 229.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27431-v30.xml emd-27431-v30.xml emd-27431.xml emd-27431.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27431_fsc.xml emd_27431_fsc.xml | 13.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_27431.png emd_27431.png | 83.9 KB | ||
| Masks |  emd_27431_msk_1.map emd_27431_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-27431.cif.gz emd-27431.cif.gz | 5.2 KB | ||
| Others |  emd_27431_additional_1.map.gz emd_27431_additional_1.map.gz emd_27431_half_map_1.map.gz emd_27431_half_map_1.map.gz emd_27431_half_map_2.map.gz emd_27431_half_map_2.map.gz | 122 MB 226.2 MB 226.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27431 http://ftp.pdbj.org/pub/emdb/structures/EMD-27431 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27431 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27431 | HTTPS FTP |
-Related structure data
| Related structure data |  8dh7MC  8dh6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27431.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27431.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map sharp | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.831 Å | ||||||||||||||||||||
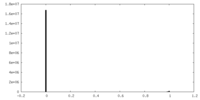
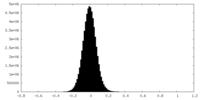
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27431_msk_1.map emd_27431_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
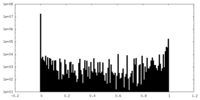
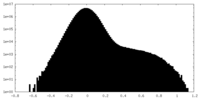
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: map
| File | emd_27431_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map | ||||||||||||
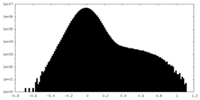
| Projections & Slices |
| ||||||||||||
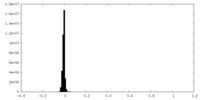
| Density Histograms |
-Half map: halfA
| File | emd_27431_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfA | ||||||||||||
| Projections & Slices |
| ||||||||||||
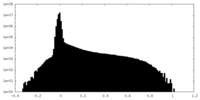
| Density Histograms |
-Half map: #1
| File | emd_27431_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Succinyl-CoA:acetate CoA-transferase
| Entire | Name: Succinyl-CoA:acetate CoA-transferase |
|---|---|
| Components |
|
-Supramolecule #1: Succinyl-CoA:acetate CoA-transferase
| Supramolecule | Name: Succinyl-CoA:acetate CoA-transferase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) |
-Macromolecule #1: Acetyl-CoA hydrolase
| Macromolecule | Name: Acetyl-CoA hydrolase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number:  acetyl-CoA hydrolase acetyl-CoA hydrolase |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) |
| Molecular weight | Theoretical: 58.791785 KDa |
| Sequence | String: MTISNLLKQR VRYAPYLKKV KEAHELIPLF KNGQYLGWSG FTGVGTPKAV PEALIDHVEK NNLQGKLRFN LFVGASAGPE ENRWAEHDM IIKRAPHQVG KPIAKAINQG RIEFFDKHLS MFPQDLTYGF YTRERKDNKI LDYTIIEATA IKEDGSIVPG P SVGGSPEF ...String: MTISNLLKQR VRYAPYLKKV KEAHELIPLF KNGQYLGWSG FTGVGTPKAV PEALIDHVEK NNLQGKLRFN LFVGASAGPE ENRWAEHDM IIKRAPHQVG KPIAKAINQG RIEFFDKHLS MFPQDLTYGF YTRERKDNKI LDYTIIEATA IKEDGSIVPG P SVGGSPEF ITVSDKVIIE VNTATPSFEG IHDIDMPVNP PFRKPYPYLK VDDKCGVDSI PVDPEKVVAI VESTMRDQVP PN TPSDDMS RAIAGHLVEF FRNEVKHGRL PENLLPLQSG IGNIANAVIE GLAGAQFKHL TVWTEVLQDS FLDLFENGSL DYA TATSVR LTEKGFDRAF ANWENFKHRL CLRSQVVSNN PEMIRRLGVI AMNTPVEVDI YAHANSTNVN GSRMLNGLGG SADF LRNAK LSIMHAPSAR PTKVDPTGIS TIVPMASHVD QTEHDLDILV TDQGLADLRG LSPKERAREI INKCAHPDYQ ALLTD YLDR AEHYAKKHNC LHEPHMLKNA FKFHTNLAEK GTMKVDSWEP VD UniProtKB:  Acetyl-CoA hydrolase Acetyl-CoA hydrolase |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.1 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 3.1 µm / Nominal defocus min: 1.0 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 0.989314222 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


 Z
Z Y
Y X
X