+ Open data
Open data
- Basic information
Basic information
| Entry | 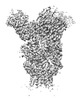 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
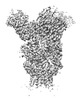
| Title | H1 Solomon Islands 2006 hemagglutinin in complex with Ab109 | |||||||||
 Map data Map data | H1 Solomon Islands 2006 in complex with ab109 | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information viral budding from plasma membrane / clathrin-dependent endocytosis of virus by host cell / host cell surface receptor binding / apical plasma membrane / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral budding from plasma membrane / clathrin-dependent endocytosis of virus by host cell / host cell surface receptor binding / apical plasma membrane / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane viral envelope / virion attachment to host cell / host cell plasma membrane / virion membraneSimilarity search - Function | |||||||||
| Biological species |   Influenza A virus (A/Solomon Islands/3/2006(H1N1)) / Influenza A virus (A/Solomon Islands/3/2006(H1N1)) /   Mus musculus (house mouse) Mus musculus (house mouse) | |||||||||
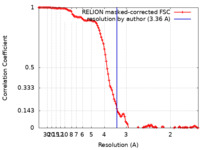
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.36 Å cryo EM / Resolution: 3.36 Å | |||||||||
 Authors Authors | Windsor IW / Caradonna TM / Schmidt AG | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2022 Journal: Cell Rep / Year: 2022Title: An epitope-enriched immunogen expands responses to a conserved viral site. Authors: Timothy M Caradonna / Larance Ronsard / Ashraf S Yousif / Ian W Windsor / Rachel Hecht / Thalia Bracamonte-Moreno / Anne A Roffler / Max J Maron / Daniel P Maurer / Jared Feldman / Elisa ...Authors: Timothy M Caradonna / Larance Ronsard / Ashraf S Yousif / Ian W Windsor / Rachel Hecht / Thalia Bracamonte-Moreno / Anne A Roffler / Max J Maron / Daniel P Maurer / Jared Feldman / Elisa Marchiori / Ralston M Barnes / Daniel Rohrer / Nils Lonberg / Thomas H Oguin / Gregory D Sempowski / Thomas B Kepler / Masayuki Kuraoka / Daniel Lingwood / Aaron G Schmidt /  Abstract: Pathogens evade host humoral responses by accumulating mutations in surface antigens. While variable, there are conserved regions that cannot mutate without compromising fitness. Antibodies targeting ...Pathogens evade host humoral responses by accumulating mutations in surface antigens. While variable, there are conserved regions that cannot mutate without compromising fitness. Antibodies targeting these conserved epitopes are often broadly protective but remain minor components of the repertoire. Rational immunogen design leverages a structural understanding of viral antigens to modulate humoral responses to favor these responses. Here, we report an epitope-enriched immunogen presenting a higher copy number of the influenza hemagglutinin (HA) receptor-binding site (RBS) epitope relative to other B cell epitopes. Immunization in a partially humanized murine model imprinted with an H1 influenza shows H1-specific serum and >99% H1-specific B cells being RBS-directed. Single B cell analyses show a genetically restricted response that structural analysis defines as RBS-directed antibodies engaging the RBS with germline-encoded contacts. These data show how epitope enrichment expands B cell responses toward conserved epitopes and advances immunogen design approaches for next-generation viral vaccines. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26605.map.gz emd_26605.map.gz | 127.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26605-v30.xml emd-26605-v30.xml emd-26605.xml emd-26605.xml | 15 KB 15 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26605_fsc.xml emd_26605_fsc.xml | 11.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_26605.png emd_26605.png | 99.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26605 http://ftp.pdbj.org/pub/emdb/structures/EMD-26605 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26605 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26605 | HTTPS FTP |
-Related structure data
| Related structure data |  7ummMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26605.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26605.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | H1 Solomon Islands 2006 in complex with ab109 | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : hemagglutinin H1 Solomon Islands/03/2006 in complex with ab109 Fab
| Entire | Name: hemagglutinin H1 Solomon Islands/03/2006 in complex with ab109 Fab |
|---|---|
| Components |
|
-Supramolecule #1: hemagglutinin H1 Solomon Islands/03/2006 in complex with ab109 Fab
| Supramolecule | Name: hemagglutinin H1 Solomon Islands/03/2006 in complex with ab109 Fab type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|
-Supramolecule #2: Hemagglutinin
| Supramolecule | Name: Hemagglutinin / type: complex / Chimera: Yes / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Influenza A virus (A/Solomon Islands/3/2006(H1N1)) Influenza A virus (A/Solomon Islands/3/2006(H1N1)) |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
-Supramolecule #3: ab109 Fab heavy chain, ab 109 Fab light chain
| Supramolecule | Name: ab109 Fab heavy chain, ab 109 Fab light chain / type: complex / Chimera: Yes / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Hemagglutinin
| Macromolecule | Name: Hemagglutinin / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Influenza A virus (A/Solomon Islands/3/2006(H1N1)) Influenza A virus (A/Solomon Islands/3/2006(H1N1))Strain: A/Solomon Islands/3/2006(H1N1) |
| Molecular weight | Theoretical: 58.608316 KDa |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: ADPGYLLEDT ICIGYHANNS TDTVDTVLEK NVTVTHSVNL LEDSHNGKLC LLKGIAPLQL GNCSVAGWIL GNPECELLIS RESWSYIVE KPNPENGTCY PGHFADYEEL REQLSSVSSF ERFEIFPKES SWPNHTTTGV SASCSHNGES SFYKNLLWLT G KNGLYPNL ...String: ADPGYLLEDT ICIGYHANNS TDTVDTVLEK NVTVTHSVNL LEDSHNGKLC LLKGIAPLQL GNCSVAGWIL GNPECELLIS RESWSYIVE KPNPENGTCY PGHFADYEEL REQLSSVSSF ERFEIFPKES SWPNHTTTGV SASCSHNGES SFYKNLLWLT G KNGLYPNL SKSYANNKEK EVLVLWGVHH PPNIGDQRAL YHTENAYVSV VSSHYSRKFT PEIAKRPKVR DQEGRINYYW TL LEPGDTI IFEANGNLIA PRYAFALSRG FGSGIINSNA PMDECDAKCQ TPQGAINSSL PFQNVHPVTI GECPKYVRSA KLR MVTGLR NIPSIQSRGL FGAIAGFIEG GWTGMVDGWY GYHHQNEQGS GYAADQKSTQ NAINGITNKV NSVIEKMNTQ FTAV GKEFN KLERRMENLN KKVDDGFIDI WTYNAELLVL LENERTLDFH DSNVKNLYEK VKSQLKNNAK EIGNGCFEFY HKCND ECME SVKNGTYDYP KYSEESKLNR EKIDGVRSGS GGALEVLFQ |
-Macromolecule #2: ab109 Fab heavy chain
| Macromolecule | Name: ab109 Fab heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Molecular weight | Theoretical: 23.267232 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGAE VKKPGASVKV SCKASGYTFT GYYIHWVRQA PGQGLEWMGW INPNTGGTVY AQTFQARVTM TRDTSISTAY MELIRLRSD DTAVYYCARE RGTGAPDAFN IWGQGTLVTV SGASTKGPSV FPLAPSGTAA LGCLVKDYFP EPVTVSWNSG A LTSGVHTF ...String: QVQLVQSGAE VKKPGASVKV SCKASGYTFT GYYIHWVRQA PGQGLEWMGW INPNTGGTVY AQTFQARVTM TRDTSISTAY MELIRLRSD DTAVYYCARE RGTGAPDAFN IWGQGTLVTV SGASTKGPSV FPLAPSGTAA LGCLVKDYFP EPVTVSWNSG A LTSGVHTF PAVLQSSGLY SLSSVVTVPS SSLGTQTYIC NVNHKPSNTK VDKKVEPKSC |
-Macromolecule #3: ab109 Fab light chain
| Macromolecule | Name: ab109 Fab light chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Molecular weight | Theoretical: 23.806414 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIVMTQSPAS LAVSLGQRAT ISCRASKSVS TSGYSYIHWY QQKPGQPPKL LIYLATNLES GVPARFSGSG SGTDFTLNIH PVEEEDAAT YYCQHSRDTP YTFGGGTKLE IKRTVAAPSV FIFPPSDEQL KSGTASVVCL LNNFYPREAK VQWKVDNALQ S GNSQESVT ...String: DIVMTQSPAS LAVSLGQRAT ISCRASKSVS TSGYSYIHWY QQKPGQPPKL LIYLATNLES GVPARFSGSG SGTDFTLNIH PVEEEDAAT YYCQHSRDTP YTFGGGTKLE IKRTVAAPSV FIFPPSDEQL KSGTASVVCL LNNFYPREAK VQWKVDNALQ S GNSQESVT EQDSKDSTYS LSSTLTLSKA DYEKHKVYAC EVTHQGLSSP VTKSFNRGEC |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 1.07 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller