[English] 日本語
 Yorodumi
Yorodumi- EMDB-26361: Structure of E. coli dGTPase bound to T7 bacteriophage protein Gp... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of E. coli dGTPase bound to T7 bacteriophage protein Gp1.2 and dGTP | ||||||||||||
 Map data Map data | DeepEMhancer post-processed map | ||||||||||||
 Sample Sample |
| ||||||||||||
| Function / homology |  Function and homology information Function and homology information dGTPase / dGTPase /  dGTPase activity / dGTP catabolic process / nucleobase-containing small molecule interconversion / cobalt ion binding / dGTPase activity / dGTP catabolic process / nucleobase-containing small molecule interconversion / cobalt ion binding /  single-stranded DNA binding / manganese ion binding / single-stranded DNA binding / manganese ion binding /  DNA replication / DNA replication /  GTPase activity / magnesium ion binding / identical protein binding GTPase activity / magnesium ion binding / identical protein bindingSimilarity search - Function | ||||||||||||
| Biological species |   Escherichia coli str. K-12 substr. MG1655 (bacteria) / Escherichia coli str. K-12 substr. MG1655 (bacteria) /    Escherichia phage T7 (virus) Escherichia phage T7 (virus) | ||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.1 Å cryo EM / Resolution: 3.1 Å | ||||||||||||
 Authors Authors | Klemm BP / Dillard LB / Borgnia MJ / Schaaper RM | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Mechanism by which T7 bacteriophage protein Gp1.2 inhibits dGTPase. Authors: Bradley P Klemm / Deepa Singh / Cassandra E Smith / Allen L Hsu / Lucas B Dillard / Juno M Krahn / Robert E London / Geoffrey A Mueller / Mario J Borgnia / Roel M Schaaper /  Abstract: Levels of the cellular dNTPs, the direct precursors for DNA synthesis, are important for DNA replication fidelity, cell cycle control, and resistance against viruses. encodes a dGTPase (2'- ...Levels of the cellular dNTPs, the direct precursors for DNA synthesis, are important for DNA replication fidelity, cell cycle control, and resistance against viruses. encodes a dGTPase (2'-deoxyguanosine-5'-triphosphate [dGTP] triphosphohydrolase [dGTPase]; gene, Dgt) that establishes the normal dGTP level required for accurate DNA replication but also plays a role in protecting against bacteriophage T7 infection by limiting the dGTP required for viral DNA replication. T7 counteracts Dgt using an inhibitor, the gene product (Gp1.2). This interaction is a useful model system for studying the ongoing evolutionary virus/host "arms race." We determined the structure of Gp1.2 by NMR spectroscopy and solved high-resolution cryo-electron microscopy structures of the Dgt-Gp1.2 complex also including either dGTP substrate or GTP coinhibitor bound in the active site. These structures reveal the mechanism by which Gp1.2 inhibits Dgt and indicate that Gp1.2 preferentially binds the GTP-bound form of Dgt. Biochemical assays reveal that the two inhibitors use different modes of inhibition and bind to Dgt in combination to yield enhanced inhibition. We thus propose an in vivo inhibition model wherein the Dgt-Gp1.2 complex equilibrates with GTP to fully inactivate Dgt, limiting dGTP hydrolysis and preserving the dGTP pool for viral DNA replication. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26361.map.gz emd_26361.map.gz | 40.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26361-v30.xml emd-26361-v30.xml emd-26361.xml emd-26361.xml | 30.1 KB 30.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26361_fsc.xml emd_26361_fsc.xml | 8.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_26361.png emd_26361.png | 169.5 KB | ||
| Masks |  emd_26361_msk_1.map emd_26361_msk_1.map | 47.6 MB |  Mask map Mask map | |
| Others |  emd_26361_additional_1.map.gz emd_26361_additional_1.map.gz emd_26361_additional_2.map.gz emd_26361_additional_2.map.gz emd_26361_additional_3.map.gz emd_26361_additional_3.map.gz emd_26361_half_map_1.map.gz emd_26361_half_map_1.map.gz emd_26361_half_map_2.map.gz emd_26361_half_map_2.map.gz | 9.2 MB 43.7 MB 36.5 MB 36.7 MB 36.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26361 http://ftp.pdbj.org/pub/emdb/structures/EMD-26361 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26361 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26361 | HTTPS FTP |
-Related structure data
| Related structure data |  7u66MC  7u65C  7u67C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26361.map.gz / Format: CCP4 / Size: 47.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26361.map.gz / Format: CCP4 / Size: 47.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer post-processed map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.932 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
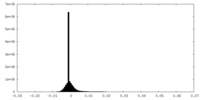
-Mask #1
| File |  emd_26361_msk_1.map emd_26361_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
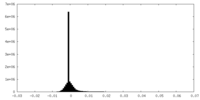
-Additional map: RELION post-processed map
| File | emd_26361_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION post-processed map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
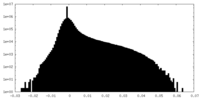
-Additional map: PHENIX auto-sharpened map
| File | emd_26361_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PHENIX auto-sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Full map from RELION refinement
| File | emd_26361_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full map from RELION refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 2
| File | emd_26361_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 1
| File | emd_26361_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : dGTPase hexamer bound to six copies of Gp1.2
| Entire | Name: dGTPase hexamer bound to six copies of Gp1.2 |
|---|---|
| Components |
|
-Supramolecule #1: dGTPase hexamer bound to six copies of Gp1.2
| Supramolecule | Name: dGTPase hexamer bound to six copies of Gp1.2 / type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Escherichia coli str. K-12 substr. MG1655 (bacteria) Escherichia coli str. K-12 substr. MG1655 (bacteria) |
-Supramolecule #2: dGTP triphosphohydrolase
| Supramolecule | Name: dGTP triphosphohydrolase / type: complex / Chimera: Yes / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Escherichia coli str. K-12 substr. MG1655 (bacteria) Escherichia coli str. K-12 substr. MG1655 (bacteria) |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) / Recombinant plasmid: pET30-Ek Escherichia coli BL21(DE3) (bacteria) / Recombinant plasmid: pET30-Ek |
-Supramolecule #3: Gene 1.2 protein
| Supramolecule | Name: Gene 1.2 protein / type: complex / Chimera: Yes / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:    Escherichia phage T7 (virus) Escherichia phage T7 (virus) |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) / Recombinant plasmid: pDest-566 Escherichia coli BL21(DE3) (bacteria) / Recombinant plasmid: pDest-566 |
-Macromolecule #1: Deoxyguanosinetriphosphate triphosphohydrolase
| Macromolecule | Name: Deoxyguanosinetriphosphate triphosphohydrolase / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number:  dGTPase dGTPase |
|---|---|
| Source (natural) | Organism:   Escherichia coli str. K-12 substr. MG1655 (bacteria) Escherichia coli str. K-12 substr. MG1655 (bacteria)Strain: K12 |
| Molecular weight | Theoretical: 59.470863 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: MAQIDFRKKI NWHRRYRSPQ GVKTEHEILR IFESDRGRII NSPAIRRLQQ KTQVFPLERN AAVRTRLTHS MEVQQVGRYI AKEILSRLK ELKLLEAYGL DELTGPFESI VEMSCLMHDI GNPPFGHFGE AAINDWFRQR LHPEDAESQP LTDDRCSVAA L RLRDGEEP ...String: MAQIDFRKKI NWHRRYRSPQ GVKTEHEILR IFESDRGRII NSPAIRRLQQ KTQVFPLERN AAVRTRLTHS MEVQQVGRYI AKEILSRLK ELKLLEAYGL DELTGPFESI VEMSCLMHDI GNPPFGHFGE AAINDWFRQR LHPEDAESQP LTDDRCSVAA L RLRDGEEP LNELRRKIRQ DLCHFEGNAQ GIRLVHTLMR MNLTWAQVGG ILKYTRPAWW RGETPETHHY LMKKPGYYLS EE AYIARLR KELNLALYSR FPLTWIMEAA DDISYCVADL EDAVEKRIFT VEQLYHHLHE AWGQHEKGSL FSLVVENAWE KSR SNSLSR STEDQFFMYL RVNTLNKLVP YAAQRFIDNL PAIFAGTFNH ALLEDASECS DLLKLYKNVA VKHVFSHPDV ERLE LQGYR VISGLLEIYR PLLSLSLSDF TELVEKERVK RFPIESRLFH KLSTRHRLAY VEAVSKLPSD SPEFPLWEYY YRCRL LQDY ISGMTDLYAW DEYRRLMAVE Q |
-Macromolecule #2: Inhibitor of dGTPase
| Macromolecule | Name: Inhibitor of dGTPase / type: protein_or_peptide / ID: 2 Details: The additional N-terminal sequence is retained after cleavage of the expression tag with TEV protease. Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Escherichia phage T7 (virus) Escherichia phage T7 (virus) |
| Molecular weight | Theoretical: 10.595868 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: GSFTMGRLYS GNLAAFKAAT NKLFQLDLAV IYDDWYDAYT RKDCIRLRIE DRSGNLIDTS TFYHHDEDVL FNMCTDWLNH MYDQLKDWK |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 6 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 6 / Formula: DGT |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-DGT: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 290 K / Instrument: LEICA EM GP | |||||||||||||||
| Details | Frozen stocks were thawed and mixed to a final concentration of 1.25:1 Gp1.2 to dGTPase (monomer) along with 1 mM dGTP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 45000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 45000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Number grids imaged: 1 / Number real images: 1206 / Average exposure time: 8.4 sec. / Average electron dose: 54.0 e/Å2 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Details | The dGTPase-Gp1.2 cryo-EM model was fit into the EM map, then the dGTP built in using Coot. |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-7u66: |
 Movie
Movie Controller
Controller






 Z
Z Y
Y X
X