+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
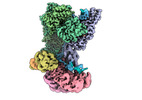
| Title | I-F3b Cascade-TniQ full R-loop complex | |||||||||
 Map data Map data | combined map of local-resolution filtered maps from full refinement and focused refinement | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  CRISPR-Cas / CRISPR-Cas /  Transposon / CAST / Cascade / I-F / I-F3 / Transposon / CAST / Cascade / I-F / I-F3 /  DNA BINDING PROTEIN / DNA BINDING PROTEIN-DNA-RNA complex DNA BINDING PROTEIN / DNA BINDING PROTEIN-DNA-RNA complex | |||||||||
| Biological species |   Aeromonas salmonicida (bacteria) Aeromonas salmonicida (bacteria) | |||||||||
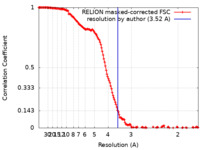
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.52 Å cryo EM / Resolution: 3.52 Å | |||||||||
 Authors Authors | Park JU / Mehrotra E / Kellogg EH | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2023 Journal: Mol Cell / Year: 2023Title: Multiple adaptations underly co-option of a CRISPR surveillance complex for RNA-guided DNA transposition. Authors: Jung-Un Park / Michael T Petassi / Shan-Chi Hsieh / Eshan Mehrotra / Gabriel Schuler / Jagat Budhathoki / Vinh H Truong / Summer B Thyme / Ailong Ke / Elizabeth H Kellogg / Joseph E Peters /  Abstract: CRISPR-associated transposons (CASTs) are natural RNA-directed transposition systems. We demonstrate that transposon protein TniQ plays a central role in promoting R-loop formation by RNA-guided DNA- ...CRISPR-associated transposons (CASTs) are natural RNA-directed transposition systems. We demonstrate that transposon protein TniQ plays a central role in promoting R-loop formation by RNA-guided DNA-targeting modules. TniQ residues, proximal to CRISPR RNA (crRNA), are required for recognizing different crRNA categories, revealing an unappreciated role of TniQ to direct transposition into different classes of crRNA targets. To investigate adaptations allowing CAST elements to utilize attachment sites inaccessible to CRISPR-Cas surveillance complexes, we compared and contrasted PAM sequence requirements in both I-F3b CAST and I-F1 CRISPR-Cas systems. We identify specific amino acids that enable a wider range of PAM sequences to be accommodated in I-F3b CAST elements compared with I-F1 CRISPR-Cas, enabling CAST elements to access attachment sites as sequences drift and evade host surveillance. Together, this evidence points to the central role of TniQ in facilitating the acquisition of CRISPR effector complexes for RNA-guided DNA transposition. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26348.map.gz emd_26348.map.gz | 7.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26348-v30.xml emd-26348-v30.xml emd-26348.xml emd-26348.xml | 31.7 KB 31.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26348_fsc.xml emd_26348_fsc.xml | 15 KB | Display |  FSC data file FSC data file |
| Images |  emd_26348.png emd_26348.png | 81.8 KB | ||
| Others |  emd_26348_additional_1.map.gz emd_26348_additional_1.map.gz emd_26348_additional_2.map.gz emd_26348_additional_2.map.gz emd_26348_additional_3.map.gz emd_26348_additional_3.map.gz emd_26348_additional_4.map.gz emd_26348_additional_4.map.gz emd_26348_half_map_1.map.gz emd_26348_half_map_1.map.gz emd_26348_half_map_2.map.gz emd_26348_half_map_2.map.gz | 225.1 MB 51.6 MB 51.9 MB 51.9 MB 225.5 MB 225.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26348 http://ftp.pdbj.org/pub/emdb/structures/EMD-26348 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26348 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26348 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26348.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26348.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | combined map of local-resolution filtered maps from full refinement and focused refinement | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.873 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: unsharpened map of the full structure
| File | emd_26348_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map of the full structure | ||||||||||||
| Projections & Slices |
| ||||||||||||
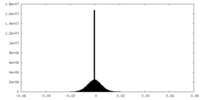
| Density Histograms |
-Additional map: Unsharpened map from focused refinement
| File | emd_26348_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map from focused refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: halfmap 1 from focused refinement
| File | emd_26348_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap 1 from focused refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
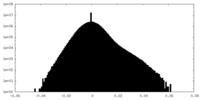
| Density Histograms |
-Additional map: halfmap 2 from focused refinement
| File | emd_26348_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap 2 from focused refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
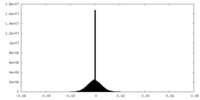
| Density Histograms |
-Half map: Halfmap 2 of the full structure
| File | emd_26348_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Halfmap 2 of the full structure | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Halfmap 1 of the full structure
| File | emd_26348_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Halfmap 1 of the full structure | ||||||||||||
| Projections & Slices |
| ||||||||||||
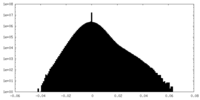
| Density Histograms |
- Sample components
Sample components
-Entire : I-F3b Cascade-TniQ Full R-loop complex
| Entire | Name: I-F3b Cascade-TniQ Full R-loop complex |
|---|---|
| Components |
|
-Supramolecule #1: I-F3b Cascade-TniQ Full R-loop complex
| Supramolecule | Name: I-F3b Cascade-TniQ Full R-loop complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Aeromonas salmonicida (bacteria) Aeromonas salmonicida (bacteria) |
| Molecular weight | Theoretical: 450 KDa |
-Macromolecule #1: crRNA
| Macromolecule | Name: crRNA / type: rna / ID: 1 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:   Aeromonas salmonicida (bacteria) Aeromonas salmonicida (bacteria) |
| Molecular weight | Theoretical: 19.399604 KDa |
| Sequence | String: CCAAGAAAAG GACUGGAAGA AAUCAUCCAA GUUGGGGACU AUUUUCUGCC GUAUAGGCAG |
-Macromolecule #2: Target strand DNA
| Macromolecule | Name: Target strand DNA / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Aeromonas salmonicida (bacteria) Aeromonas salmonicida (bacteria) |
| Molecular weight | Theoretical: 35.61368 KDa |
| Sequence | String: (DC)(DT)(DG)(DG)(DC)(DT)(DG)(DG)(DC)(DG) (DA)(DA)(DC)(DG)(DA)(DG)(DC)(DG)(DC)(DA) (DA)(DG)(DG)(DT)(DG)(DG)(DT)(DG)(DG) (DC)(DC)(DC)(DC)(DA)(DT)(DC)(DA)(DG)(DC) (DC) (DA)(DC)(DA)(DT)(DC)(DC) ...String: (DC)(DT)(DG)(DG)(DC)(DT)(DG)(DG)(DC)(DG) (DA)(DA)(DC)(DG)(DA)(DG)(DC)(DG)(DC)(DA) (DA)(DG)(DG)(DT)(DG)(DG)(DT)(DG)(DG) (DC)(DC)(DC)(DC)(DA)(DT)(DC)(DA)(DG)(DC) (DC) (DA)(DC)(DA)(DT)(DC)(DC)(DC)(DG) (DG)(DC)(DA)(DC)(DT)(DC)(DG)(DA)(DA)(DG) (DT)(DC) (DC)(DC)(DC)(DA)(DA)(DC)(DT) (DT)(DG)(DG)(DA)(DT)(DG)(DA)(DT)(DT)(DT) (DC)(DT)(DT) (DC)(DC)(DA)(DG)(DT)(DC) (DC)(DT)(DG)(DG)(DT)(DA)(DA)(DG)(DC)(DA) (DC)(DC)(DC)(DG) (DA)(DA)(DT)(DC)(DA) (DT)(DC)(DC)(DT)(DC)(DT)(DT)(DG)(DC)(DG) (DG) |
-Macromolecule #3: Non-target strand DNA
| Macromolecule | Name: Non-target strand DNA / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Aeromonas salmonicida (bacteria) Aeromonas salmonicida (bacteria) |
| Molecular weight | Theoretical: 35.737723 KDa |
| Sequence | String: (DC)(DC)(DG)(DC)(DA)(DA)(DG)(DA)(DG)(DG) (DA)(DT)(DG)(DA)(DT)(DT)(DC)(DG)(DG)(DG) (DT)(DG)(DC)(DT)(DT)(DA)(DC)(DC)(DT) (DC)(DC)(DT)(DG)(DA)(DC)(DC)(DT)(DT)(DC) (DT) (DT)(DT)(DA)(DG)(DT)(DA) ...String: (DC)(DC)(DG)(DC)(DA)(DA)(DG)(DA)(DG)(DG) (DA)(DT)(DG)(DA)(DT)(DT)(DC)(DG)(DG)(DG) (DT)(DG)(DC)(DT)(DT)(DA)(DC)(DC)(DT) (DC)(DC)(DT)(DG)(DA)(DC)(DC)(DT)(DT)(DC) (DT) (DT)(DT)(DA)(DG)(DT)(DA)(DG)(DG) (DT)(DT)(DC)(DA)(DA)(DC)(DC)(DC)(DC)(DT) (DG)(DA) (DT)(DC)(DG)(DA)(DG)(DT)(DG) (DC)(DC)(DG)(DG)(DG)(DA)(DT)(DG)(DT)(DG) (DG)(DC)(DT) (DG)(DA)(DT)(DG)(DG)(DG) (DG)(DC)(DC)(DA)(DC)(DC)(DA)(DC)(DC)(DT) (DT)(DG)(DC)(DG) (DC)(DT)(DC)(DG)(DT) (DT)(DC)(DG)(DC)(DC)(DA)(DG)(DC)(DC)(DA) (DG) |
-Macromolecule #4: Cas8/5
| Macromolecule | Name: Cas8/5 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Aeromonas salmonicida (bacteria) Aeromonas salmonicida (bacteria) |
| Molecular weight | Theoretical: 78.728367 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MVTIMHIEEL LDIEDHGERD RQLRRYLAPY SAEIGVDGAE KMALVVLLNL TLKRDRVESL CDEGLARQLL SDEGHITNCL HTVRWLHTH NLKYPDARVS GERLIINAPP LIPGVISSAG LPMRMGWAHD SSDINLAKLF GTSFRYRDDS TNLALQLVAR S KTWEQALI ...String: MVTIMHIEEL LDIEDHGERD RQLRRYLAPY SAEIGVDGAE KMALVVLLNL TLKRDRVESL CDEGLARQLL SDEGHITNCL HTVRWLHTH NLKYPDARVS GERLIINAPP LIPGVISSAG LPMRMGWAHD SSDINLAKLF GTSFRYRDDS TNLALQLVAR S KTWEQALI GLGLTQQQLD IWCQLLASNL ENNTFPTVVS PFSKQVRFLY QGNYCVVTPV VSHALLAQLQ NVVHEKKLQC TY IHHDHPA SVGSLVGALG GKVAVLDYPP PVSPDKARSF SQARKHRLAN GQSLFDRSVF NDHVFIDALK HVISRPGLTR KQQ RQLRLS ALRYLRRQLA IWLGPIIEWR DEIVSSGRGE PGNLPSGGLE LELITQPKKM LPELMLQVAG RFHLELQNHS AGRR FAFHP ALMAPIKSQI LWLLRQLADD EEKDEPHPPT SCYYLHLSGL TVYDASALAN PYLCGIPSLS ALAGFCHDYE RRLQS LIGQ SVYFRGLAWY LGRYSLVTGK HLPEPSKSAD PKSVSAIRRP GLLDGRYCDL GMDLIIEVHI PTGGSLPFTT CLDLLR VAL PARFAGGCLH PPSLYEEYNW CTVYQDKSTL FTVLSRLPRY GCWIYPSDAD LRSFEELSEA LALDRRLRPV ATGFVFL EE PVERAGSIEG QHVYAESAIG TALCINPVEM RLAGKKRFFG AGFWQLNDAK GAILMNGSAN TG |
-Macromolecule #5: Cas7
| Macromolecule | Name: Cas7 / type: protein_or_peptide / ID: 5 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Aeromonas salmonicida (bacteria) Aeromonas salmonicida (bacteria) |
| Molecular weight | Theoretical: 39.105172 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MELCTHLSYS RSLSPGKAVF FYKTAESDFV PLRIEVAKIS GQKCGYTEGF DANLKPKNIE RYELAYSNPQ TIEACYVPPN VDELYCRFS LRVEANSMRP YVCSNPDVLR VMIGLAQAYQ RLGGYNELAR RYSANVLRGI WLWRNQYTQG TKIEIKTSLG S TYHIPDAR ...String: MELCTHLSYS RSLSPGKAVF FYKTAESDFV PLRIEVAKIS GQKCGYTEGF DANLKPKNIE RYELAYSNPQ TIEACYVPPN VDELYCRFS LRVEANSMRP YVCSNPDVLR VMIGLAQAYQ RLGGYNELAR RYSANVLRGI WLWRNQYTQG TKIEIKTSLG S TYHIPDAR RLSWSGDWPE LEQKQLEQLT SEMAKALSQP DIFWFADVTA SLKTGFCQEI FPSQKFTERP DDHSVASRQL AT VECSDGQ LAACINPQKI GAALQKIDDW WANDADLPLR VHEYGANHEA LTALRHPATG QDFYHLLTKA EQFVTVLESS EGG GVELPG EVHYLMAVLV KGGLFQKGKG R |
-Macromolecule #6: Cas6
| Macromolecule | Name: Cas6 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Aeromonas salmonicida (bacteria) Aeromonas salmonicida (bacteria) |
| Molecular weight | Theoretical: 23.640832 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MTENRYFFAI RYLSDDVDCG LLAGRCISIL HGFRQAHPGI QIGVAFPEWS DRDLGRSIAF VSTNKSLLER FRERSYFQVM QADNFFALS LVLEVPDTCQ NVRFIRNQNL AKLFVGERRR RLARAKRRAK ARGEAFQPHM PDETKVVGVF HSVFMQSASS G QSYILHIQ ...String: MTENRYFFAI RYLSDDVDCG LLAGRCISIL HGFRQAHPGI QIGVAFPEWS DRDLGRSIAF VSTNKSLLER FRERSYFQVM QADNFFALS LVLEVPDTCQ NVRFIRNQNL AKLFVGERRR RLARAKRRAK ARGEAFQPHM PDETKVVGVF HSVFMQSASS G QSYILHIQ KHRYERSEDS GYSSYGLASN DLYTGYVPDL GAIFSTLF |
-Macromolecule #7: TniQ
| Macromolecule | Name: TniQ / type: protein_or_peptide / ID: 7 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Aeromonas salmonicida (bacteria) Aeromonas salmonicida (bacteria) |
| Molecular weight | Theoretical: 46.366637 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MHLLVRPEPF ADEALESYFL RLSQENGFER YRIFSGSVQD WLHTTDHAAA GAFPLELSRL NIFHASRSSG LRVRALQLVD RLTDGAPFR LLQLALCHSA ISFGNHYKAV HRSGVDIPLS FIRVHQIPCC PDCLRESAYV RQCWHFKPYV GCHRHGGRLI Y SCPACGES ...String: MHLLVRPEPF ADEALESYFL RLSQENGFER YRIFSGSVQD WLHTTDHAAA GAFPLELSRL NIFHASRSSG LRVRALQLVD RLTDGAPFR LLQLALCHSA ISFGNHYKAV HRSGVDIPLS FIRVHQIPCC PDCLRESAYV RQCWHFKPYV GCHRHGGRLI Y SCPACGES LNYLASESIN HCQCGFDLRT ASTVPAQPDE IQLSALAYGC SFESSNPLLA IGSLSARFGA LYWYQQRYLS DH EAVRDDR ALTKAIGHFT AWPDAFWREL QQMVDDALVR QTKPLNHTDF VDVFGSVVAD CRQIPMRNTG QNFILKNLIG FLT DLVARH PQCRVANVGD LLLSAVDAAT LLSTSVEQVR RLHHEGFLPL SIRPASRNTV SPHRAVFHLR HVVELRQARM QSHH DHSST YLPAW |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.9 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)