+ Open data
Open data
- Basic information
Basic information
| Entry | 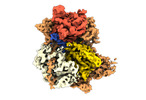 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
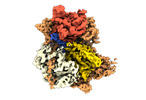
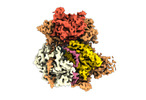
| Title | Structure of DrmAB:ADP:DNA complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |   Serratia (bacteria) / synthetic construct (others) Serratia (bacteria) / synthetic construct (others) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.4 Å cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Bravo JPK / Taylor DW / Brounds SJJ / Aparicio-Maldonado C | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural basis for broad anti-phage immunity by DISARM. Authors: Jack P K Bravo / Cristian Aparicio-Maldonado / Franklin L Nobrega / Stan J J Brouns / David W Taylor /    Abstract: In the evolutionary arms race against phage, bacteria have assembled a diverse arsenal of antiviral immune strategies. While the recently discovered DISARM (Defense Island System Associated with ...In the evolutionary arms race against phage, bacteria have assembled a diverse arsenal of antiviral immune strategies. While the recently discovered DISARM (Defense Island System Associated with Restriction-Modification) systems can provide protection against a wide range of phage, the molecular mechanisms that underpin broad antiviral targeting but avoiding autoimmunity remain enigmatic. Here, we report cryo-EM structures of the core DISARM complex, DrmAB, both alone and in complex with an unmethylated phage DNA mimetic. These structures reveal that DrmAB core complex is autoinhibited by a trigger loop (TL) within DrmA and binding to DNA substrates containing a 5' overhang dislodges the TL, initiating a long-range structural rearrangement for DrmAB activation. Together with structure-guided in vivo studies, our work provides insights into the mechanism of phage DNA recognition and specific activation of this widespread antiviral defense system. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24939.map.gz emd_24939.map.gz | 117.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24939-v30.xml emd-24939-v30.xml emd-24939.xml emd-24939.xml | 14 KB 14 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24939.png emd_24939.png | 80.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24939 http://ftp.pdbj.org/pub/emdb/structures/EMD-24939 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24939 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24939 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24939.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24939.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : DrmAB:ADP:DNA
| Entire | Name: DrmAB:ADP:DNA |
|---|---|
| Components |
|
-Supramolecule #1: DrmAB:ADP:DNA
| Supramolecule | Name: DrmAB:ADP:DNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|
-Supramolecule #2: DrmAB
| Supramolecule | Name: DrmAB / type: complex / Chimera: Yes / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Serratia (bacteria) Serratia (bacteria) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Supramolecule #3: DNA
| Supramolecule | Name: DNA / type: complex / Chimera: Yes / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: DrmA
| Macromolecule | Name: DrmA / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Serratia (bacteria) Serratia (bacteria) |
| Molecular weight | Theoretical: 147.980766 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MTDNNKSSKT PNAWIAIDHN FSRAQVLTYY TLQLKGEHSL HQAISDNDWI LVLDTTGNIT RVGRILRIRS DLETTTIFFD RMLQVKSVV SIGITPFKFP PNDRAGRIQW TDFIETLPKE LHITIADIPK IEDQTYIREL LQLAVMDDLL GPAGGPNELI V DMGVRDRY ...String: MTDNNKSSKT PNAWIAIDHN FSRAQVLTYY TLQLKGEHSL HQAISDNDWI LVLDTTGNIT RVGRILRIRS DLETTTIFFD RMLQVKSVV SIGITPFKFP PNDRAGRIQW TDFIETLPKE LHITIADIPK IEDQTYIREL LQLAVMDDLL GPAGGPNELI V DMGVRDRY LVGKLAPREA AERGQEFPID AEDIEDEEPD LIVKAKTAKV NSPSVLGSGE TDTAEEIDAA SNQSLVPSSL GM TFCVDGD VDRVEIEARW GRYERVPNDE HQFFKSNGQK AKVWKRIPCG GKIVLPLIEG SISHNAPDST SPEVRVQGSI RAK NDNGDR LITLFLVNAQ EEPDTNRDTA WVFQPELIVR AAKDAAKPAI FRRRPVLDAD GMDPEREALE MIYRDRVEFA VGHG VAVHA EIADDVTLAT EVRTTVMPQY EVQATETPGL ELSDRPAMRE MVSSGLLDMQ RLATLDIDPL VDALSVLTND YATWI DEQN LNVSSKAKGF DTQAQTAINR CQEIHTRLQE GINTLKSNEN ALAAFRFANQ AMATQRIRSL YALAMRRGED VTLDKF DVL KNRSWRPFQL AFLLLSIPSL ADPCHPDRVK PIEAYADLLW FPTGGGKTEA YLGVAAFTMA IRRMQGNLGG YDSSRGL TV IMRYTLRLLT LQQFQRATAL ICAMEVLRRE ALNKGDKSLG TEPFTIGLWV GNKVTPGTTE DSHNAIEKTR NPGSYNAG T ASPVQLTSCP WCGTEIVPGQ DVEVKKDKAG GRTFVYCGDK KGRCEFSKGK SSTQPHPGIP VLVVDEEIYH RPPTMMIAT VDKFAMMAWR GQVRTLFGRV EKECERHGLL WPGANCTGNH QAFKGQPSAK VKAIPPIRPP DLIIQDEFHL ISGPLGTMVG LYETAVDEL CSWTLNGKTV KPKIIASTAT VRKAKEQVNN VFMRQVSVFP PHGLDVEDNF FSVQRHIKDK FGRRYLGVCS P GSSRPAML IRVYTAFLTA AQELFDHFGE PADPYMTMVG YFNSLRELGG MKRLAEDDVQ TRSYRVQMSM VERPALAQRS VN NIRELTS RVSSQDIPKY LDNLEVKFKA EFDSSAGKYV TKWQEGDTRA IDVVLATNML SVGVDVNRLG LMAVNGQPKG TAE YIQATS RVGRSFPGLV CTVLTWARPR DLSHYETFEH YHATFYKHVE AQSVTPFSPR AMDRGLTGSL LSLMRLKNNE FSPN EGAGK LDMSNQSELA HAIEVLATRA GNVAEDNARK LLAENELKER ADEWAKEASK GGRILGYEKR GPDKDKTVAL IKSPG LQAW DNWTVPMSMR EVESGVRLIM DTKFIKDDHD WKPRPATKDE D |
-Macromolecule #2: DrmB
| Macromolecule | Name: DrmB / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Serratia (bacteria) Serratia (bacteria) |
| Molecular weight | Theoretical: 68.667039 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MIINNKTPVG EVRPSQLLWT YGPGALIDLP SLSVVTLGID RWERERCQPI QEARLLAAVR KVLGPQVENL RMPPFQKSEL VDPWSAEAN IGVPVRPFPR WMRCVKCGLL SPFDDGLLEI KEDRFRAERT RFVHKGCTGS KGNLPAKDAD AVPARFLLAC R DGHLDDFP ...String: MIINNKTPVG EVRPSQLLWT YGPGALIDLP SLSVVTLGID RWERERCQPI QEARLLAAVR KVLGPQVENL RMPPFQKSEL VDPWSAEAN IGVPVRPFPR WMRCVKCGLL SPFDDGLLEI KEDRFRAERT RFVHKGCTGS KGNLPAKDAD AVPARFLLAC R DGHLDDFP WHYFVHGGNS TCKGTLRFFE SGASLQTENL WVRCDSCEAS RSMAHAFGKA GKENLPACRG RHPHLDQFDI DC GEEPRAV LLGATNSWFP ITLSALAIPQ SKNPLSQLIQ DGWPLFEAIT AEVMVPIVVQ TLKLTGGLPG IDKYSVSDIW SAI EMHRSG GDSEFVGEAD IKGPEWEVLT EANPPTDYPH FMSKKIGTPA QFIPYISRVL LLERLREVNA LLGFTRVEAP EGSG EINER PQMASLARNK PEWVPANQVH GEGIFIQFNE KTLVAWESLD AVKQVDEMLR GGHTGWRNSR NLDPNEDYPG IRYAM LHTL SHLLIRELAL ECGYNAASIR ERIYADTSNG SPQAGILIYT AAADSDGTLG GLVDLGKPEN LGRLLVQALN RSKICS SDP LCSEHNPEKD RSLHAAACHA CTLVAETSCE QGNRYLDRSL LIPTLERIHA AFFKGF |
-Macromolecule #3: DNA (5'-D(P*TP*TP*TP*TP*TP*TP*T)-3')
| Macromolecule | Name: DNA (5'-D(P*TP*TP*TP*TP*TP*TP*T)-3') / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 2.084392 KDa |
| Sequence | String: (DT)(DT)(DT)(DT)(DT)(DT)(DT) |
-Macromolecule #4: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 1 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 150 mM NaCl, 25 mM HEPES pH 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 80.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
|---|---|
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 121764 |
 Movie
Movie Controller
Controller