[English] 日本語
 Yorodumi
Yorodumi- EMDB-22974: Structure of DPP9 bound to catalytically-inactive CARD8 (S297A) -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22974 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of DPP9 bound to catalytically-inactive CARD8 (S297A) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationCARD8 inflammasome complex assembly /  NACHT domain binding / Formation of apoptosome / cysteine-type endopeptidase activator activity / inhibition of cysteine-type endopeptidase activity / negative regulation of NLRP3 inflammasome complex assembly / NACHT domain binding / Formation of apoptosome / cysteine-type endopeptidase activator activity / inhibition of cysteine-type endopeptidase activity / negative regulation of NLRP3 inflammasome complex assembly /  NLRP3 inflammasome complex / NLRP3 inflammasome complex /  dipeptidyl-peptidase IV / dipeptidyl-peptidase IV /  CARD domain binding / negative regulation of lipopolysaccharide-mediated signaling pathway ...CARD8 inflammasome complex assembly / CARD domain binding / negative regulation of lipopolysaccharide-mediated signaling pathway ...CARD8 inflammasome complex assembly /  NACHT domain binding / Formation of apoptosome / cysteine-type endopeptidase activator activity / inhibition of cysteine-type endopeptidase activity / negative regulation of NLRP3 inflammasome complex assembly / NACHT domain binding / Formation of apoptosome / cysteine-type endopeptidase activator activity / inhibition of cysteine-type endopeptidase activity / negative regulation of NLRP3 inflammasome complex assembly /  NLRP3 inflammasome complex / NLRP3 inflammasome complex /  dipeptidyl-peptidase IV / dipeptidyl-peptidase IV /  CARD domain binding / negative regulation of lipopolysaccharide-mediated signaling pathway / self proteolysis / dipeptidyl-peptidase activity / Regulation of the apoptosome activity / negative regulation of programmed cell death / CARD domain binding / negative regulation of lipopolysaccharide-mediated signaling pathway / self proteolysis / dipeptidyl-peptidase activity / Regulation of the apoptosome activity / negative regulation of programmed cell death /  Hydrolases; Acting on peptide bonds (peptidases) / regulation of canonical NF-kappaB signal transduction / Hydrolases; Acting on peptide bonds (peptidases) / regulation of canonical NF-kappaB signal transduction /  pattern recognition receptor activity / negative regulation of interleukin-1 beta production / negative regulation of NF-kappaB transcription factor activity / pyroptotic inflammatory response / cell leading edge / cysteine-type endopeptidase activator activity involved in apoptotic process / antiviral innate immune response / positive regulation of cysteine-type endopeptidase activity involved in apoptotic process / negative regulation of tumor necrosis factor-mediated signaling pathway / negative regulation of canonical NF-kappaB signal transduction / pattern recognition receptor activity / negative regulation of interleukin-1 beta production / negative regulation of NF-kappaB transcription factor activity / pyroptotic inflammatory response / cell leading edge / cysteine-type endopeptidase activator activity involved in apoptotic process / antiviral innate immune response / positive regulation of cysteine-type endopeptidase activity involved in apoptotic process / negative regulation of tumor necrosis factor-mediated signaling pathway / negative regulation of canonical NF-kappaB signal transduction /  aminopeptidase activity / serine-type peptidase activity / molecular condensate scaffold activity / positive regulation of interleukin-1 beta production / aminopeptidase activity / serine-type peptidase activity / molecular condensate scaffold activity / positive regulation of interleukin-1 beta production /  peptidase activity / regulation of apoptotic process / defense response to virus / peptidase activity / regulation of apoptotic process / defense response to virus /  microtubule / protein homodimerization activity / protein-containing complex / microtubule / protein homodimerization activity / protein-containing complex /  proteolysis / proteolysis /  nucleoplasm / identical protein binding / nucleoplasm / identical protein binding /  nucleus / nucleus /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
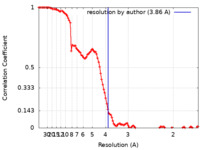
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.86 Å cryo EM / Resolution: 3.86 Å | |||||||||
 Authors Authors | Hollingsworth LR / Sharif H / Wu H | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Immunity / Year: 2021 Journal: Immunity / Year: 2021Title: Dipeptidyl peptidase 9 sets a threshold for CARD8 inflammasome formation by sequestering its active C-terminal fragment. Authors: Humayun Sharif / L Robert Hollingsworth / Andrew R Griswold / Jeffrey C Hsiao / Qinghui Wang / Daniel A Bachovchin / Hao Wu /  Abstract: CARD8 detects intracellular danger signals and forms a caspase-1 activating inflammasome. Like the related inflammasome sensor NLRP1, CARD8 autoprocesses into noncovalently associated N-terminal (NT) ...CARD8 detects intracellular danger signals and forms a caspase-1 activating inflammasome. Like the related inflammasome sensor NLRP1, CARD8 autoprocesses into noncovalently associated N-terminal (NT) and C-terminal (CT) fragments and binds the cellular dipeptidyl peptidases DPP8 and 9 (DPP8/9). Certain danger-associated signals, including the DPP8/9 inhibitor Val-boroPro (VbP) and HIV protease, induce proteasome-mediated NT degradation and thereby liberate the inflammasome-forming CT. Here, we report cryoelectron microscopy (cryo-EM) structures of CARD8 bound to DPP9, revealing a repressive ternary complex consisting of DPP9, full-length CARD8, and CARD8-CT. Unlike NLRP1-CT, CARD8-CT does not interact with the DPP8/9 active site and is not directly displaced by VbP. However, larger DPP8/9 active-site probes can directly weaken this complex in vitro, and VbP itself nevertheless appears to disrupt this complex, perhaps indirectly, in cells. Thus, DPP8/9 inhibitors can activate the CARD8 inflammasome by promoting CARD8 NT degradation and by weakening ternary complex stability. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22974.map.gz emd_22974.map.gz | 62.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22974-v30.xml emd-22974-v30.xml emd-22974.xml emd-22974.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22974_fsc.xml emd_22974_fsc.xml | 11.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_22974.png emd_22974.png | 140.7 KB | ||
| Others |  emd_22974_additional_1.map.gz emd_22974_additional_1.map.gz | 118.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22974 http://ftp.pdbj.org/pub/emdb/structures/EMD-22974 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22974 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22974 | HTTPS FTP |
-Related structure data
| Related structure data |  7jkqC  7jn7C C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10600 (Title: Human DPP9-CARD8 complex-S297A / Data size: 967.9 EMPIAR-10600 (Title: Human DPP9-CARD8 complex-S297A / Data size: 967.9 Data #1: Unaligned multi frame micographs of CARD8-DPP9-S297A-TILT [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22974.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22974.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Sharpened Map
| File | emd_22974_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened Map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : DPP9 bound to catalytically-inactive CARD8 (S297A)
| Entire | Name: DPP9 bound to catalytically-inactive CARD8 (S297A) |
|---|---|
| Components |
|
-Supramolecule #1: DPP9 bound to catalytically-inactive CARD8 (S297A)
| Supramolecule | Name: DPP9 bound to catalytically-inactive CARD8 (S297A) / type: cell / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: DPP9
| Macromolecule | Name: DPP9 / type: other / ID: 1 / Classification: other |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GAMGSMATTG TPTADRGDAA ATDDPAARFQ VQKHSWDGLR SIIHGSRKYS GLIVNKAPHD FQFVQKTDES GPHSHRLYYL GMPYGSRENS LLYSEIPKKV RKEALLLLSW KQMLDHFQAT PHHGVYSREE ELLRERKRLG VFGITSYDFH SESGLFLFQA SNSLFHCRDG ...String: GAMGSMATTG TPTADRGDAA ATDDPAARFQ VQKHSWDGLR SIIHGSRKYS GLIVNKAPHD FQFVQKTDES GPHSHRLYYL GMPYGSRENS LLYSEIPKKV RKEALLLLSW KQMLDHFQAT PHHGVYSREE ELLRERKRLG VFGITSYDFH SESGLFLFQA SNSLFHCRDG GKNGFMVSPM KPLEIKTQCS GPRMDPKICP ADPAFFSFIN NSDLWVANIE TGEERRLTFC HQGLSNVLDD PKSAGVATFV IQEEFDRFTG YWWCPTASWE GSEGLKTLRI LYEEVDESEV EVIHVPSPAL EERKTDSYRY PRTGSKNPKI ALKLAEFQTD SQGKIVSTQE KELVQPFSSL FPKVEYIARA GWTRDGKYAW AMFLDRPQQW LQLVLLPPAL FIPSTENEEQ RLASARAVPR NVQPYVVYEE VTNVWINVHD IFYPFPQSEG EDELCFLRAN ECKTGFCHLY KVTAVLKSQG YDWSEPFSPG EDEFKCPIKE EIALTSGEWE VLARHGSKIW VNEETKLVYF QGTKDTPLEH HLYVVSYEAA GEIVRLTTPG FSHSCSMSQN FDMFVSHYSS VSTPPCVHVY KLSGPDDDPL HKQPRFWASM MEAASCPPDY VPPEIFHFHT RSDVRLYGMI YKPHALQPGK KHPTVLFVYG GPQVQLVNNS FKGIKYLRLN TLASLGYAVV VIDGRGSCQR GLRFEGALKN QMGQVEIEDQ VEGLQFVAEK YGFIDLSRVA IHGWSYGGFL SLMGLIHKPQ VFKVAIAGAP VTVWMAYDTG YTERYMDVPE NNQHGYEAGS VALHVEKLPN EPNRLLILHG FLDENVHFFH TNFLVSQLIR AGKPYQLQIY PNERHSIRCP ESGEHYEVTL LHFLQEYL |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #2: CARD8
| Macromolecule | Name: CARD8 / type: other / ID: 2 / Classification: other |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEKKECPEKS SSSEEELPRR DSGSSRNIDA SKLIRLQGSR KLLVDNSIRE LQYTKTGIFF QAEACVTNDT VYRELPCVSE TLCDISHFFQ EDDETEAEPL LFRAVPECQL SGGDIPSVSE EQESSEGQDS GDICSEENQI VSSYASKVCF EIEEDYKNRQ FLGPEGNVDV ...String: MEKKECPEKS SSSEEELPRR DSGSSRNIDA SKLIRLQGSR KLLVDNSIRE LQYTKTGIFF QAEACVTNDT VYRELPCVSE TLCDISHFFQ EDDETEAEPL LFRAVPECQL SGGDIPSVSE EQESSEGQDS GDICSEENQI VSSYASKVCF EIEEDYKNRQ FLGPEGNVDV ELIDKSTNRY SVWFPTAGWY LWSATGLGFL VRDEVTVTIA FGSWSQHLAL DLQHHEQWLV GGPLFDVTAE PEEAVAEIHL PHFISLQAGE VDVSWFLVAH FKNEGMVLEH PARVEPFYAV LESPSFALMG ILLRIASGTR LSIPITSNTL IYYHPHPEDI KFHLYLVPSD ALLTKAIDDE EDRFHGVRLQ TSPPMEPLNF GSSYIVSNSA NLKVMPKELK LSYRSPGEIQ HFSKFYAGQM KEPIQLEITE KRHGTLVWDT EVKPVDLQLV AASAPPPFSG AAFVKENHRQ LQARMGDLKG VLDDLQDNEV LTENEKELVE QEKTRQSKNE ALLSMVEKKG DLALDVLFRS ISERDPYLVS YLRQQNL |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.40 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: Crosslinking with glutaraldehyde was done for 5-10 minutes on ice immediately before plunging grids. | |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: -0.0025 µm / Nominal defocus min: -0.0008 µm / Nominal magnification: 105000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: -0.0025 µm / Nominal defocus min: -0.0008 µm / Nominal magnification: 105000 |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 3840 / Average exposure time: 1.5 sec. / Average electron dose: 63.67 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z
Z Y
Y X
X