+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21661 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Hexameric NanR-DNA hetero-complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NanR dimer-DNA hetero-complex /  transcriptional regulator / GntR superfamily / transcriptional regulator / GntR superfamily /  sialic acid / sialic acid /  Neu5Ac / Neu5Ac /  cooperativity / cooperativity /  Gene regulation. / Gene regulation. /  GENE REGULATION GENE REGULATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationDNA-binding transcription factor activity / negative regulation of DNA-templated transcription /  DNA binding DNA bindingSimilarity search - Function | |||||||||
| Biological species |   Escherichia coli (E. coli) / synthetic construct (others) Escherichia coli (E. coli) / synthetic construct (others) | |||||||||
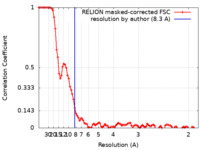
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 8.3 Å cryo EM / Resolution: 8.3 Å | |||||||||
 Authors Authors | Hariprasad V / Horne C | |||||||||
| Funding support |  New Zealand, 1 items New Zealand, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Mechanism of NanR gene repression and allosteric induction of bacterial sialic acid metabolism. Authors: Christopher R Horne / Hariprasad Venugopal / Santosh Panjikar / David M Wood / Amy Henrickson / Emre Brookes / Rachel A North / James M Murphy / Rosmarie Friemann / Michael D W Griffin / ...Authors: Christopher R Horne / Hariprasad Venugopal / Santosh Panjikar / David M Wood / Amy Henrickson / Emre Brookes / Rachel A North / James M Murphy / Rosmarie Friemann / Michael D W Griffin / Georg Ramm / Borries Demeler / Renwick C J Dobson /      Abstract: Bacteria respond to environmental changes by inducing transcription of some genes and repressing others. Sialic acids, which coat human cell surfaces, are a nutrient source for pathogenic and ...Bacteria respond to environmental changes by inducing transcription of some genes and repressing others. Sialic acids, which coat human cell surfaces, are a nutrient source for pathogenic and commensal bacteria. The Escherichia coli GntR-type transcriptional repressor, NanR, regulates sialic acid metabolism, but the mechanism is unclear. Here, we demonstrate that three NanR dimers bind a (GGTATA)-repeat operator cooperatively and with high affinity. Single-particle cryo-electron microscopy structures reveal the DNA-binding domain is reorganized to engage DNA, while three dimers assemble in close proximity across the (GGTATA)-repeat operator. Such an interaction allows cooperative protein-protein interactions between NanR dimers via their N-terminal extensions. The effector, N-acetylneuraminate, binds NanR and attenuates the NanR-DNA interaction. The crystal structure of NanR in complex with N-acetylneuraminate reveals a domain rearrangement upon N-acetylneuraminate binding to lock NanR in a conformation that weakens DNA binding. Our data provide a molecular basis for the regulation of bacterial sialic acid metabolism. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21661.map.gz emd_21661.map.gz | 35.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21661-v30.xml emd-21661-v30.xml emd-21661.xml emd-21661.xml | 18.4 KB 18.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21661_fsc.xml emd_21661_fsc.xml | 9.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_21661.png emd_21661.png | 78.8 KB | ||
| Masks |  emd_21661_msk_1.map emd_21661_msk_1.map | 70.2 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-21661.cif.gz emd-21661.cif.gz | 6.2 KB | ||
| Others |  emd_21661_half_map_1.map.gz emd_21661_half_map_1.map.gz emd_21661_half_map_2.map.gz emd_21661_half_map_2.map.gz | 65.2 MB 65.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21661 http://ftp.pdbj.org/pub/emdb/structures/EMD-21661 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21661 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21661 | HTTPS FTP |
-Related structure data
| Related structure data |  6wg7MC  6wfqC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_21661.map.gz / Format: CCP4 / Size: 70.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21661.map.gz / Format: CCP4 / Size: 70.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.94 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_21661_msk_1.map emd_21661_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_21661_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_21661_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Hexameric NanR-DNA hetero-complex
| Entire | Name: Hexameric NanR-DNA hetero-complex |
|---|---|
| Components |
|
-Supramolecule #1: Hexameric NanR-DNA hetero-complex
| Supramolecule | Name: Hexameric NanR-DNA hetero-complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Hexameric NanR-DNA hetero-complex |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Molecular weight | Theoretical: 198.5 KDa |
-Macromolecule #1: DNA (35-MER)
| Macromolecule | Name: DNA (35-MER) / type: dna / ID: 1 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 10.856016 KDa |
| Sequence | String: (DT)(DT)(DG)(DA)(DT)(DC)(DT)(DG)(DG)(DT) (DA)(DT)(DA)(DA)(DC)(DA)(DG)(DG)(DT)(DA) (DT)(DA)(DA)(DA)(DG)(DG)(DT)(DA)(DT) (DA)(DT)(DC)(DG)(DT)(DT) |
-Macromolecule #2: DNA (35-MER)
| Macromolecule | Name: DNA (35-MER) / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 10.673923 KDa |
| Sequence | String: (DA)(DA)(DC)(DG)(DA)(DT)(DA)(DT)(DA)(DC) (DC)(DT)(DT)(DT)(DA)(DT)(DA)(DC)(DC)(DT) (DG)(DT)(DT)(DA)(DT)(DA)(DC)(DC)(DA) (DG)(DA)(DT)(DC)(DA)(DA) |
-Macromolecule #3: HTH-type transcriptional repressor NanR
| Macromolecule | Name: HTH-type transcriptional repressor NanR / type: protein_or_peptide / ID: 3 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Molecular weight | Theoretical: 29.566354 KDa |
| Recombinant expression | Organism:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) |
| Sequence | String: MGLMNAFDSQ TEDSSPAIGR NLRSRPLARK KLSEMVEEEL EQMIRRREFG EGEQLPSERE LMAFFNVGRP SVREALAALK RKGLVQINN GERARVSRPS ADTIIGELSG MAKDFLSHPG GIAHFEQLRL FFESSLVRYA AEHATDEQID LLAKALEINS Q SLDNNAAF ...String: MGLMNAFDSQ TEDSSPAIGR NLRSRPLARK KLSEMVEEEL EQMIRRREFG EGEQLPSERE LMAFFNVGRP SVREALAALK RKGLVQINN GERARVSRPS ADTIIGELSG MAKDFLSHPG GIAHFEQLRL FFESSLVRYA AEHATDEQID LLAKALEINS Q SLDNNAAF IRSDVDFHRV LAEIPGNPIF MAIHVALLDW LIAARPTVTD QALHEHNNVS YQQHIAIVDA IRRHDPDEAD RA LQSHLNS VSATWHAFGQ TTNKKK UniProtKB: HTH-type transcriptional repressor NanR |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 20mM Tris-HCL,50mM NaCl, pH 8.0 |
| Grid | Model: UltrAuFoil / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK III / Details: Blot force: -3 Blot time: 3 sec Drain time: 0. |
- Electron microscopy
Electron microscopy
| Microscope | TFS TALOS |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm Bright-field microscopy / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 2287 / Average exposure time: 51.54 sec. / Average electron dose: 45.0 e/Å2 |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Protocol: RIGID BODY FIT | ||||||
| Output model |  PDB-6wg7: |
 Movie
Movie Controller
Controller













 Z
Z Y
Y X
X