[English] 日本語
 Yorodumi
Yorodumi- EMDB-21156: Cryo-EM Structure of Escherichia coli 2-oxoglutarate dehydrogenas... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21156 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of Escherichia coli 2-oxoglutarate dehydrogenase E1 component sucA | |||||||||
 Map data Map data | Escherichia coli 2-oxoglutarate dehydrogenase E1 component sucA | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TCA cycle / 2-oxoglutarate dehydrogenase complex (OGDH) / E1 component / sucA / AMP / Oxaloacetate (OAA) / Dimer /  OXIDOREDUCTASE OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology information oxoglutarate dehydrogenase (succinyl-transferring) / oxoglutarate dehydrogenase (succinyl-transferring) /  oxoglutarate dehydrogenase (succinyl-transferring) activity / oxoglutarate dehydrogenase (succinyl-transferring) activity /  oxoglutarate dehydrogenase complex / oxoglutarate dehydrogenase complex /  thiamine pyrophosphate binding / thiamine pyrophosphate binding /  tricarboxylic acid cycle / tricarboxylic acid cycle /  nucleotide binding / magnesium ion binding / identical protein binding / nucleotide binding / magnesium ion binding / identical protein binding /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Escherichia coli K-12 (bacteria) Escherichia coli K-12 (bacteria) | |||||||||
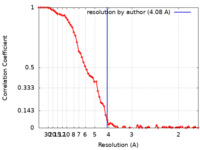
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.08 Å cryo EM / Resolution: 4.08 Å | |||||||||
 Authors Authors | Gao H | |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM Structure of Escherichia coli 2-oxoglutarate dehydrogenase E1 component sucA Authors: Gao H | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21156.map.gz emd_21156.map.gz | 28.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21156-v30.xml emd-21156-v30.xml emd-21156.xml emd-21156.xml | 11.6 KB 11.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21156_fsc.xml emd_21156_fsc.xml | 9.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_21156.png emd_21156.png | 158.5 KB | ||
| Filedesc metadata |  emd-21156.cif.gz emd-21156.cif.gz | 5.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21156 http://ftp.pdbj.org/pub/emdb/structures/EMD-21156 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21156 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21156 | HTTPS FTP |
-Related structure data
| Related structure data |  6vefMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21156.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21156.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Escherichia coli 2-oxoglutarate dehydrogenase E1 component sucA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.877 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Escherichia coli 2-oxoglutarate dehydrogenase E1 component sucA
| Entire | Name: Escherichia coli 2-oxoglutarate dehydrogenase E1 component sucA |
|---|---|
| Components |
|
-Supramolecule #1: Escherichia coli 2-oxoglutarate dehydrogenase E1 component sucA
| Supramolecule | Name: Escherichia coli 2-oxoglutarate dehydrogenase E1 component sucA type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Escherichia coli K-12 (bacteria) Escherichia coli K-12 (bacteria) |
-Macromolecule #1: 2-oxoglutarate dehydrogenase E1 component
| Macromolecule | Name: 2-oxoglutarate dehydrogenase E1 component / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO EC number:  oxoglutarate dehydrogenase (succinyl-transferring) oxoglutarate dehydrogenase (succinyl-transferring) |
|---|---|
| Source (natural) | Organism:   Escherichia coli K-12 (bacteria) / Strain: K12 Escherichia coli K-12 (bacteria) / Strain: K12 |
| Molecular weight | Theoretical: 94.942508 KDa |
| Sequence | String: DTNVKQVKVL QLINAYRFRG HQHANLDPLG LYQQDKVADL DPSFHDLTEA DFQETFNVGS FASGKETMKL GELLEALKQT YCGPIGAEY MHITSTEEKR WIQQRIESGR ATFNSEEKKR FLSELTAAEG LERYLGAKFP GAKRFSLEGD ALIPMLKEMI R HAGNSGTR ...String: DTNVKQVKVL QLINAYRFRG HQHANLDPLG LYQQDKVADL DPSFHDLTEA DFQETFNVGS FASGKETMKL GELLEALKQT YCGPIGAEY MHITSTEEKR WIQQRIESGR ATFNSEEKKR FLSELTAAEG LERYLGAKFP GAKRFSLEGD ALIPMLKEMI R HAGNSGTR EVVLGMAHRG RLNVLVNVLG KKPQDLFDEF AGKHKEHLGT GDVKYHMGFS SDFQTDGGLV HLALAFNPSH LE IVSPVVI GSVRARLDRL DEPSSNKVLP ITIHGDAAVT GQGVVQETLN MSKARGYEVG GTVRIVINNQ VGFTTSNPLD ARS TPYMTD IGKMVQAPIF HVNADDPEAV AFVTRLALDF RNTFKRDVFI DLVCYRRHGH NEADEPSATQ KIKKHPTPRK IYAD KLEQE KVATLEDATE MVNLYRDALD AGDCVVAEWR PMNMHSFTWS PYLNHEWDEE YPNKVEMKRL QELAKRISTV PEAVE MQSR VAKIYGDRQA MAAGEKLFDW GGAENLAYAT LVDEGIPVRL SGEDSGRGTF FHRHAVIHNQ SNGSTYTPLQ HIHNGQ GAF RVWDSVLSEE AVLAFEYGYA TAEPRTLTIW EAQFGDFANG AQVVIDQFIS SGEQKWGRMC GLVMLLPHGY EGQGPEH SS ARLERYLQLC AEQNMQVCVP STPAQVYHML RRQALRGMRR PLVVMSPKSL LRHPLAVSSL EELANGTFLP AIGEIDEL D PKGVKRVVMC SGKVYYDLLE QRRKNNQHDV AIVRIEQLYP FPHKAMQEVL QQFAHVKDFV WCQEEPLNQG AWYCSQHHF REVIPFGASL RYAGRPASAS PAVGHMSVHQ KQQQDLVNDA LNVE UniProtKB:  Oxoglutarate dehydrogenase (succinyl-transferring) Oxoglutarate dehydrogenase (succinyl-transferring) |
-Macromolecule #2: ADENOSINE MONOPHOSPHATE
| Macromolecule | Name: ADENOSINE MONOPHOSPHATE / type: ligand / ID: 2 / Number of copies: 2 / Formula: AMP |
|---|---|
| Molecular weight | Theoretical: 347.221 Da |
| Chemical component information |  ChemComp-AMP: |
-Macromolecule #3: OXALOACETATE ION
| Macromolecule | Name: OXALOACETATE ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: OAA |
|---|---|
| Molecular weight | Theoretical: 131.064 Da |
| Chemical component information |  ChemComp-OAA: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 80 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm Bright-field microscopy / Cs: 2.7 mm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 800 / Average exposure time: 2.0 sec. / Average electron dose: 80.0 e/Å2 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller