+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20269 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
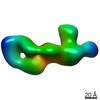
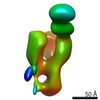
| Title | Chikungunya virus p62/E1 in complex with Fab from IgG DC2.271B | |||||||||
 Map data Map data | Chikungunya virus p62/E1 in complex with Fab from IgG DC2.271B | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /    Chikungunya virus Chikungunya virus | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 24.0 Å negative staining / Resolution: 24.0 Å | |||||||||
 Authors Authors | Pallesen J / Ward AB | |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2019 Journal: PLoS Pathog / Year: 2019Title: Human monoclonal antibodies against chikungunya virus target multiple distinct epitopes in the E1 and E2 glycoproteins. Authors: Jose A Quiroz / Ryan J Malonis / Larissa B Thackray / Courtney A Cohen / Jesper Pallesen / Rohit K Jangra / Rebecca S Brown / Daniel Hofmann / Frederick W Holtsberg / Sergey Shulenin / ...Authors: Jose A Quiroz / Ryan J Malonis / Larissa B Thackray / Courtney A Cohen / Jesper Pallesen / Rohit K Jangra / Rebecca S Brown / Daniel Hofmann / Frederick W Holtsberg / Sergey Shulenin / Elisabeth K Nyakatura / Lorellin A Durnell / Vinayak Rayannavar / Johanna P Daily / Andrew B Ward / M Javad Aman / John M Dye / Kartik Chandran / Michael S Diamond / Margaret Kielian / Jonathan R Lai /  Abstract: Chikungunya virus (CHIKV) is a mosquito-transmitted alphavirus that causes persistent arthritis in a subset of human patients. We report the isolation and functional characterization of monoclonal ...Chikungunya virus (CHIKV) is a mosquito-transmitted alphavirus that causes persistent arthritis in a subset of human patients. We report the isolation and functional characterization of monoclonal antibodies (mAbs) from two patients infected with CHIKV in the Dominican Republic. Single B cell sorting yielded a panel of 46 human mAbs of diverse germline lineages that targeted epitopes within the E1 or E2 glycoproteins. MAbs that recognized either E1 or E2 proteins exhibited neutralizing activity. Viral escape mutations localized the binding epitopes for two E1 mAbs to sites within domain I or the linker between domains I and III; and for two E2 mAbs between the β-connector region and the B-domain. Two of the E2-specific mAbs conferred protection in vivo in a stringent lethal challenge mouse model of CHIKV infection, whereas the E1 mAbs did not. These results provide insight into human antibody response to CHIKV and identify candidate mAbs for therapeutic intervention. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20269.map.gz emd_20269.map.gz | 7.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20269-v30.xml emd-20269-v30.xml emd-20269.xml emd-20269.xml | 14.9 KB 14.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20269_fsc.xml emd_20269_fsc.xml | 4.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_20269.png emd_20269.png | 53.8 KB | ||
| Others |  emd_20269_half_map_1.map.gz emd_20269_half_map_1.map.gz emd_20269_half_map_2.map.gz emd_20269_half_map_2.map.gz | 6 MB 6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20269 http://ftp.pdbj.org/pub/emdb/structures/EMD-20269 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20269 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20269 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20269.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20269.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Chikungunya virus p62/E1 in complex with Fab from IgG DC2.271B | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Chikungunya virus p62/E1 in complex with Fab from...
| File | emd_20269_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Chikungunya virus p62/E1 in complex with Fab from IgG DC2.271B, half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Chikungunya virus p62/E1 in complex with Fab from...
| File | emd_20269_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Chikungunya virus p62/E1 in complex with Fab from IgG DC2.271B, half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Chikungunya virus p62/E1 in complex with Fab from IgG DC2.271B
| Entire | Name: Chikungunya virus p62/E1 in complex with Fab from IgG DC2.271B |
|---|---|
| Components |
|
-Supramolecule #1: Chikungunya virus p62/E1 in complex with Fab from IgG DC2.271B
| Supramolecule | Name: Chikungunya virus p62/E1 in complex with Fab from IgG DC2.271B type: complex / ID: 1 / Parent: 0 |
|---|---|
| Molecular weight | Theoretical: 85 KDa |
-Supramolecule #2: Fab from IgG DC2.271B
| Supramolecule | Name: Fab from IgG DC2.271B / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Chikungunya virus p62/E1
| Supramolecule | Name: Chikungunya virus p62/E1 / type: complex / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism:    Chikungunya virus Chikungunya virus |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Staining | Type: NEGATIVE / Material: uranyl formate |
| Grid | Details: unspecified |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 57000 Bright-field microscopy / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 57000 |
| Image recording | Film or detector model: FEI CETA (4k x 4k) / Average electron dose: 30.0 e/Å2 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z
Z Y
Y X
X


















