[English] 日本語
 Yorodumi
Yorodumi- EMDB-20260: HIV Env 16055 NFL TD 2CC+ in complex with antibody 1C2 fragment a... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20260 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
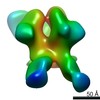
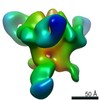
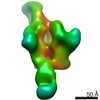
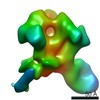
| Title | HIV Env 16055 NFL TD 2CC+ in complex with antibody 1C2 fragment antigen binding | |||||||||||||||
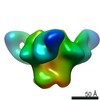
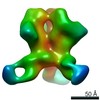
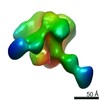
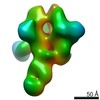
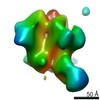
 Map data Map data | sharpened map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / virus-mediated perturbation of host defense response / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell /  viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope ...positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / virus-mediated perturbation of host defense response / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral envelope ...positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / virus-mediated perturbation of host defense response / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell /  viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity /  plasma membrane plasma membraneSimilarity search - Function | |||||||||||||||
| Biological species |    Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /   Oryctolagus cuniculus (rabbit) Oryctolagus cuniculus (rabbit) | |||||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.94 Å cryo EM / Resolution: 3.94 Å | |||||||||||||||
 Authors Authors | Ozorowski G / Torres JL / Ward AB | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Immunity / Year: 2019 Journal: Immunity / Year: 2019Title: Vaccination with Glycan-Modified HIV NFL Envelope Trimer-Liposomes Elicits Broadly Neutralizing Antibodies to Multiple Sites of Vulnerability. Authors: Viktoriya Dubrovskaya / Karen Tran / Gabriel Ozorowski / Javier Guenaga / Richard Wilson / Shridhar Bale / Christopher A Cottrell / Hannah L Turner / Gemma Seabright / Sijy O'Dell / Jonathan ...Authors: Viktoriya Dubrovskaya / Karen Tran / Gabriel Ozorowski / Javier Guenaga / Richard Wilson / Shridhar Bale / Christopher A Cottrell / Hannah L Turner / Gemma Seabright / Sijy O'Dell / Jonathan L Torres / Lifei Yang / Yu Feng / Daniel P Leaman / Néstor Vázquez Bernat / Tyler Liban / Mark Louder / Krisha McKee / Robert T Bailer / Arlette Movsesyan / Nicole A Doria-Rose / Marie Pancera / Gunilla B Karlsson Hedestam / Michael B Zwick / Max Crispin / John R Mascola / Andrew B Ward / Richard T Wyatt /    Abstract: The elicitation of broadly neutralizing antibodies (bNAbs) against the HIV-1 envelope glycoprotein (Env) trimer remains a major vaccine challenge. Most cross-conserved protein determinants are ...The elicitation of broadly neutralizing antibodies (bNAbs) against the HIV-1 envelope glycoprotein (Env) trimer remains a major vaccine challenge. Most cross-conserved protein determinants are occluded by self-N-glycan shielding, limiting B cell recognition of the underlying polypeptide surface. The exceptions to the contiguous glycan shield include the conserved receptor CD4 binding site (CD4bs) and glycoprotein (gp)41 elements proximal to the furin cleavage site. Accordingly, we performed heterologous trimer-liposome prime:boosting in rabbits to drive B cells specific for cross-conserved sites. To preferentially expose the CD4bs to B cells, we eliminated proximal N-glycans while maintaining the native-like state of the cleavage-independent NFL trimers, followed by gradual N-glycan restoration coupled with heterologous boosting. This approach successfully elicited CD4bs-directed, cross-neutralizing Abs, including one targeting a unique glycan-protein epitope and a bNAb (87% breadth) directed to the gp120:gp41 interface, both resolved by high-resolution cryoelectron microscopy. This study provides proof-of-principle immunogenicity toward eliciting bNAbs by vaccination. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20260.map.gz emd_20260.map.gz | 85.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20260-v30.xml emd-20260-v30.xml emd-20260.xml emd-20260.xml | 23 KB 23 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20260_fsc.xml emd_20260_fsc.xml | 10.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_20260.png emd_20260.png | 182.1 KB | ||
| Masks |  emd_20260_msk_1.map emd_20260_msk_1.map | 91.1 MB |  Mask map Mask map | |
| Others |  emd_20260_half_map_1.map.gz emd_20260_half_map_1.map.gz emd_20260_half_map_2.map.gz emd_20260_half_map_2.map.gz | 71.3 MB 71.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20260 http://ftp.pdbj.org/pub/emdb/structures/EMD-20260 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20260 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20260 | HTTPS FTP |
-Related structure data
| Related structure data |  6p65MC  6p62C  6pehC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20260.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20260.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_20260_msk_1.map emd_20260_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_20260_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_20260_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : HIV Env 16055 NFL TD 2CC+ in complex with antibody 1C2 fragment a...
| Entire | Name: HIV Env 16055 NFL TD 2CC+ in complex with antibody 1C2 fragment antigen binding |
|---|---|
| Components |
|
-Supramolecule #1: HIV Env 16055 NFL TD 2CC+ in complex with antibody 1C2 fragment a...
| Supramolecule | Name: HIV Env 16055 NFL TD 2CC+ in complex with antibody 1C2 fragment antigen binding type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:    Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 570 KDa |
-Macromolecule #1: HIV Env 16055 NFL TD 2CC+
| Macromolecule | Name: HIV Env 16055 NFL TD 2CC+ / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 75.630539 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPMGSLQPLA TLYLLGMLVA SVLANGNLWV TVYYGVPVWK DAETTLFCAS DAKAYEKEKH NVWATHACVP TDPNPQEMVL ENVTENFNM WKNDMVEQMH TDVISLWDQS LKPCVKLTPL CVTLECRQVN TTNATSSVNV TNGEEIKNCS FNATTELRDK K QKVYALFY ...String: MPMGSLQPLA TLYLLGMLVA SVLANGNLWV TVYYGVPVWK DAETTLFCAS DAKAYEKEKH NVWATHACVP TDPNPQEMVL ENVTENFNM WKNDMVEQMH TDVISLWDQS LKPCVKLTPL CVTLECRQVN TTNATSSVNV TNGEEIKNCS FNATTELRDK K QKVYALFY RLDIVPLEEE RKGNSSKYRL INCNTSACTQ ACPKVTFDPI PIHYCAPAGY AILKCNNKTF NGTGPCNNVS TV QCTHGIK PVVSTQLLLN GSLAEGEIII RSENLTNNVK TIIVHLNESV EIVCTRPNNY TRKSIRIGPG QTFYATGDII GNI RQAYCN ISKDDWIRTL QRVGKKLAEH FPRRIINFTS PAGGDLEITT HSFNCRGEFF YCNTSSLFNS TYNPNDTNSN SSSS NSSLD ITIPCRIKQI INMWQRVGQC MYAPPIEGNI TCKSNITGLL LVRDGGVESN ETEIFRPGGG DMRNNWRSEL YKYKV VEIK PLGIAPTRCK RRVVEGGGGS GGGGSDDDDK AVGLGAVRRG FLGAAGSTMG AASITLTVQA RQLLSGIVQQ QSNLLK APE AQQHLLQLGV WGIKQLQTRV LAIERYLKDQ QLLGIWGCSG KLICTTAVPW NSSWSNKSHD EIWGNMTWMQ WDREIGN YT NTIYRLLEDS QNQQEQNEKD LLACDGGGGS HHHHHHHH |
-Macromolecule #2: Rabbit antibody 1C2 heavy chain fragment antigen binding
| Macromolecule | Name: Rabbit antibody 1C2 heavy chain fragment antigen binding type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Oryctolagus cuniculus (rabbit) Oryctolagus cuniculus (rabbit) |
| Molecular weight | Theoretical: 27.476072 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MYRMQLLSCI ALSLALVTNS QCQSLEESGG DLVKPGASLT LTCTASGFSF GWNDYMSWVR QAPGKGLEWI GCIYAGSTRS TYYANWAKG RLTISKTSST AVTLQMTSLT AADTATYFCA RGAVTYDGLG GAYLKHFNLW GPGTLVTVSS GQPKAPSVFP L APCCGDTP ...String: MYRMQLLSCI ALSLALVTNS QCQSLEESGG DLVKPGASLT LTCTASGFSF GWNDYMSWVR QAPGKGLEWI GCIYAGSTRS TYYANWAKG RLTISKTSST AVTLQMTSLT AADTATYFCA RGAVTYDGLG GAYLKHFNLW GPGTLVTVSS GQPKAPSVFP L APCCGDTP SSTVTLGCLV KGYLPEPVTV TWNSGTLTNG VRTFPSVRQS SGLYSLSSVV SVTSSSQPVT CNVAHPATNT KV DKTVAPS TCSKHHHHHH HH |
-Macromolecule #3: Rabbit antibody 1C2 kappa chain
| Macromolecule | Name: Rabbit antibody 1C2 kappa chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Oryctolagus cuniculus (rabbit) Oryctolagus cuniculus (rabbit) |
| Molecular weight | Theoretical: 25.041023 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MYRMQLLSCI ALSLALVTNS AIKMTQTPSS VSAAVGGTVT VNCRASEDIE SYLAWYQQKP GQPPKLLIYD TSKLASGVPS RFKGSGSGT QFALTISGVQ CDDAATYYCL YGYISSDRID FGFGGGTELV VKGDPVAPSV LIFPPAADQV ATGTVTIVCV A NKYFPDVT ...String: MYRMQLLSCI ALSLALVTNS AIKMTQTPSS VSAAVGGTVT VNCRASEDIE SYLAWYQQKP GQPPKLLIYD TSKLASGVPS RFKGSGSGT QFALTISGVQ CDDAATYYCL YGYISSDRID FGFGGGTELV VKGDPVAPSV LIFPPAADQV ATGTVTIVCV A NKYFPDVT VTWEVDGTTQ TTGIENSKTP QNSADCTYNL SSTLTLTSTQ YNSHKEYTCK VTQGTTSVVQ SFNRGDC |
-Macromolecule #8: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 8 / Number of copies: 21 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: DDM added to sample shortly (< 5 minutes) before vitrification | ||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.7 µm / Nominal magnification: 36000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.7 µm / Nominal magnification: 36000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 1690 / Average exposure time: 12.5 sec. / Average electron dose: 48.0 e/Å2 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z
Z Y
Y X
X



























