[English] 日本語
 Yorodumi
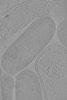
Yorodumi- EMDB-19375: Cryo-electron tomogram of Yersinia entomophaga chi2-sfGFP cells -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-electron tomogram of Yersinia entomophaga chi2-sfGFP cells | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Tc toxin /  TOXIN TOXIN | |||||||||
| Biological species |   Yersinia entomophaga (bacteria) Yersinia entomophaga (bacteria) | |||||||||
| Method |  electron tomography / electron tomography /  cryo EM cryo EM | |||||||||
 Authors Authors | Feldmueller M / Pilhofer M | |||||||||
| Funding support | European Union,  Switzerland, 2 items Switzerland, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2024 Journal: Nat Microbiol / Year: 2024Title: Stepwise assembly and release of Tc toxins from Yersinia entomophaga. Authors: Miki Feldmüller / Charles F Ericson / Pavel Afanasyev / Yun-Wei Lien / Gregor L Weiss / Florian Wollweber / Marion Schoof / Mark Hurst / Martin Pilhofer /   Abstract: Tc toxins are virulence factors of bacterial pathogens. Although their structure and intoxication mechanism are well understood, it remains elusive where this large macromolecular complex is ...Tc toxins are virulence factors of bacterial pathogens. Although their structure and intoxication mechanism are well understood, it remains elusive where this large macromolecular complex is assembled and how it is released. Here we show by an integrative multiscale imaging approach that Yersinia entomophaga Tc (YenTc) toxin components are expressed only in a subpopulation of cells that are 'primed' with several other potential virulence factors, including filaments of the protease M66/StcE. A phage-like lysis cassette is required for YenTc release; however, before resulting in complete cell lysis, the lysis cassette generates intermediate 'ghost' cells, which may serve as assembly compartments and become packed with assembled YenTc holotoxins. We hypothesize that this stepwise mechanism evolved to minimize the number of cells that need to be killed. The occurrence of similar lysis cassettes in diverse organisms indicates a conserved mechanism for Tc toxin release that may apply to other extracellular macromolecular machines. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19375.map.gz emd_19375.map.gz | 214.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19375-v30.xml emd-19375-v30.xml emd-19375.xml emd-19375.xml | 8.1 KB 8.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_19375.png emd_19375.png | 159 KB | ||
| Filedesc metadata |  emd-19375.cif.gz emd-19375.cif.gz | 3.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19375 http://ftp.pdbj.org/pub/emdb/structures/EMD-19375 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19375 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19375 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_19375.map.gz / Format: CCP4 / Size: 1.4 GB / Type: IMAGE STORED AS SIGNED INTEGER (2 BYTES) Download / File: emd_19375.map.gz / Format: CCP4 / Size: 1.4 GB / Type: IMAGE STORED AS SIGNED INTEGER (2 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 18.04 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Yersinia entomophaga chi2-sfGFP
| Entire | Name: Yersinia entomophaga chi2-sfGFP |
|---|---|
| Components |
|
-Supramolecule #1: Yersinia entomophaga chi2-sfGFP
| Supramolecule | Name: Yersinia entomophaga chi2-sfGFP / type: cell / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:   Yersinia entomophaga (bacteria) Yersinia entomophaga (bacteria) |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  electron tomography electron tomography |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
| Sectioning | Focused ion beam - Instrument: OTHER / Focused ion beam - Ion: OTHER / Focused ion beam - Voltage: 30 / Focused ion beam - Current: 0.05 / Focused ion beam - Duration: 1740 / Focused ion beam - Temperature: 123 K / Focused ion beam - Initial thickness: 2000 / Focused ion beam - Final thickness: 200 Focused ion beam - Details: The value given for _em_focused_ion_beam.instrument is Zeiss Crossbeam 550. This is not in a list of allowed values {'OTHER', 'DB235'} so OTHER is written into the XML file. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 8.0 µm / Nominal defocus min: 8.0 µm Bright-field microscopy / Nominal defocus max: 8.0 µm / Nominal defocus min: 8.0 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 2.46 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Number images used: 61 |
|---|
 Movie
Movie Controller
Controller